Structure of a NHEJ polymerase-mediated DNA synaptic complex
Brissett, N.C., Pitcher, R.S., Juarez, R., Picher, A.J., Green, A.J., Dafforn, T.R., Fox, G.C., Blanco, L., Doherty, A.J.(2007) Science 318: 456-459
- PubMed: 17947582
- DOI: https://doi.org/10.1126/science.1145112
- Primary Citation of Related Structures:
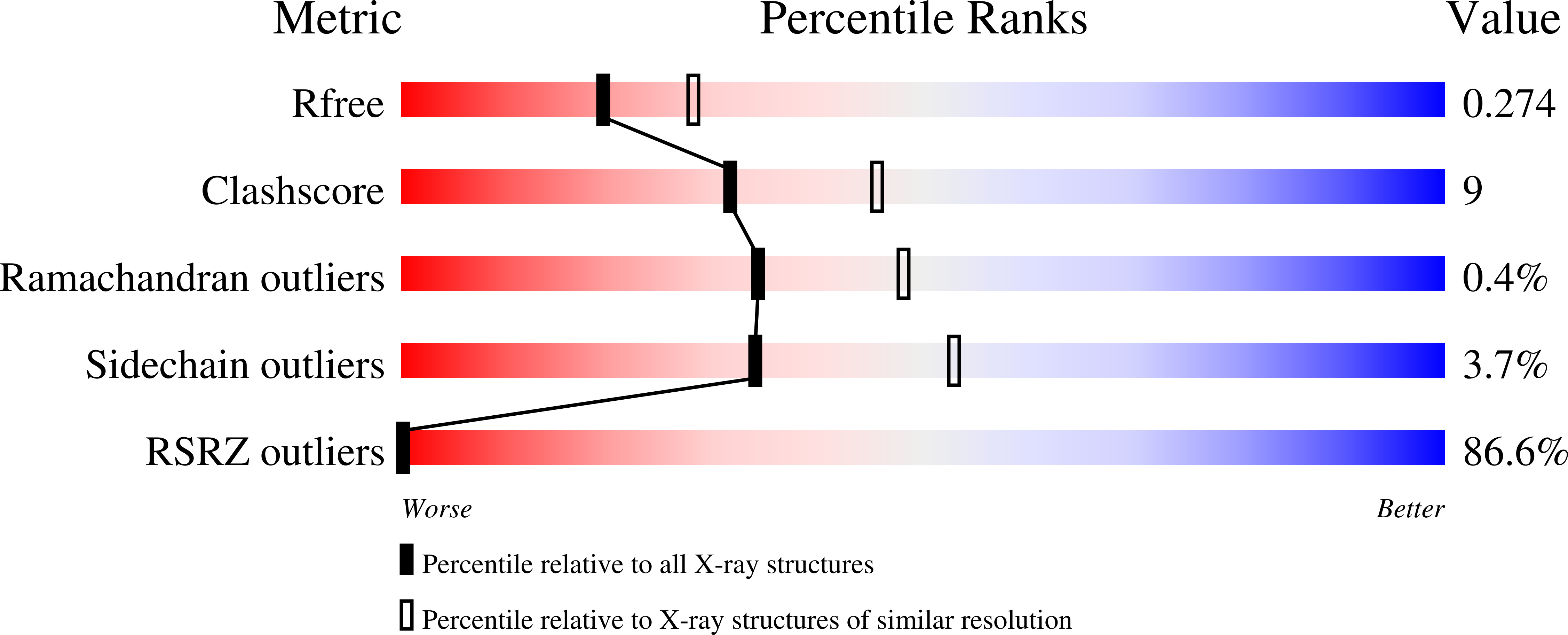
2R9L - PubMed Abstract:
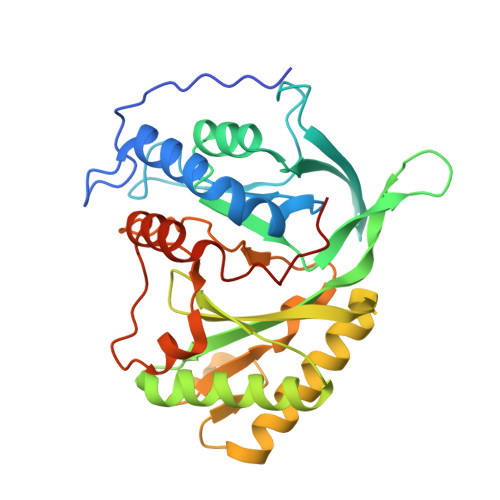
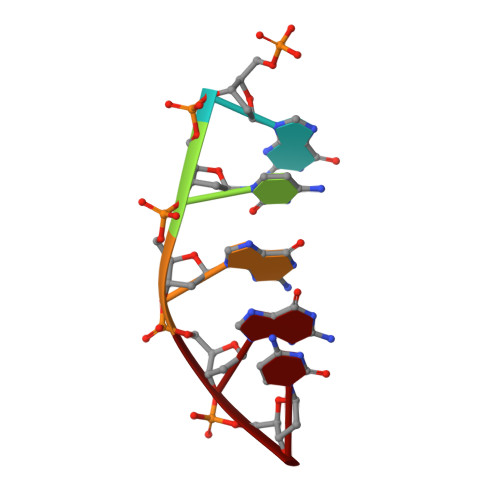
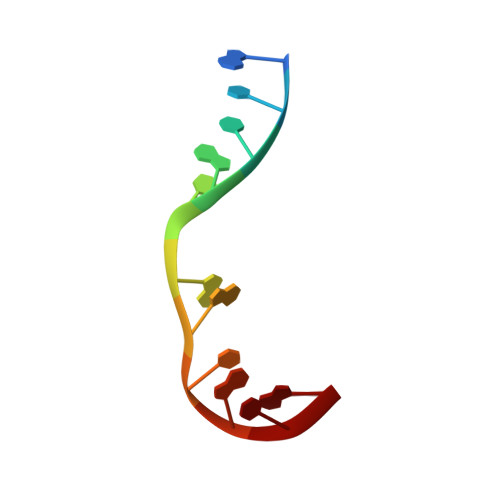
Nonhomologous end joining (NHEJ) is a critical DNA double-strand break (DSB) repair pathway required to maintain genome stability. Many prokaryotes possess a minimalist NHEJ apparatus required to repair DSBs during stationary phase, composed of two conserved core proteins, Ku and ligase D (LigD). The crystal structure of Mycobacterium tuberculosis polymerase domain of LigD mediating the synapsis of two noncomplementary DNA ends revealed a variety of interactions, including microhomology base pairing, mismatched and flipped-out bases, and 3' termini forming hairpin-like ends. Biochemical and biophysical studies confirmed that polymerase-induced end synapsis also occurs in solution. We propose that this DNA synaptic structure reflects an intermediate bridging stage of the NHEJ process, before end processing and ligation, with both the polymerase and the DNA sequence playing pivotal roles in determining the sequential order of synapsis and remodeling before end joining.
Organizational Affiliation:
Genome Damage and Stability Centre, University of Sussex, Brighton BN1 9RQ, UK.