Structural basis for substrate specificity and MSMEG_0435-0436 binding by the mycobacterial long-chain acyl-CoA carboxylase complex
Liang, Y., Rubinstein, J.L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biotin-dependent acyl-coenzyme A carboxylase alpha3 subunit | 598 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 EC: 6.3.4.14 | 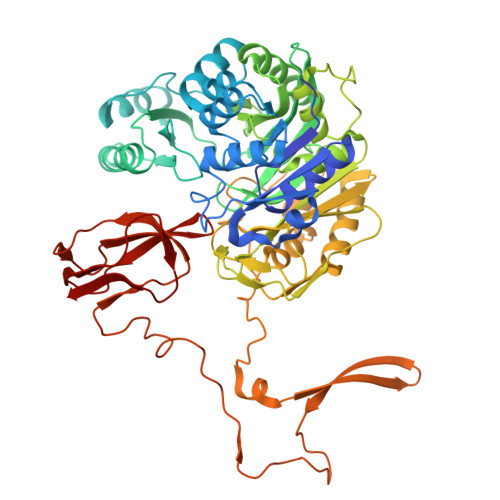 | |
UniProt | |||||
Find proteins for A0QTE1 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0QTE1 Go to UniProtKB: A0QTE1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QTE1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetyl-/propionyl-coenzyme A carboxylase AccE5 | B [auth E], M [auth P] | 94 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 | 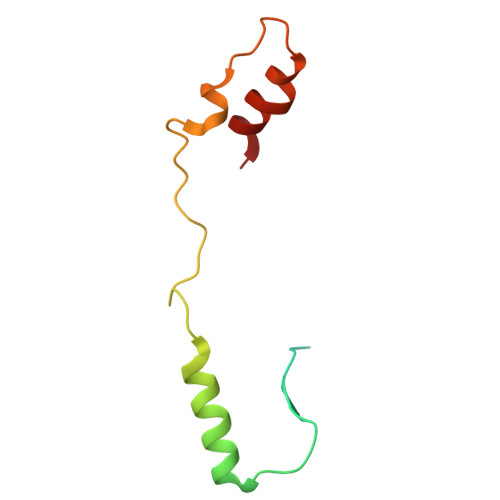 |
UniProt | |||||
Find proteins for A0QTE6 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0QTE6 Go to UniProtKB: A0QTE6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QTE6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Propionyl-CoA carboxylase beta chain | C [auth F], L [auth O] | 517 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 EC: 6.4.1.3 | 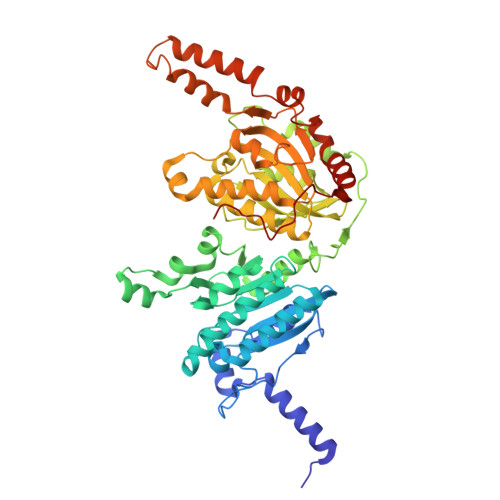 |
UniProt | |||||
Find proteins for A0R616 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0R616 Go to UniProtKB: A0R616 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0R616 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Propionyl-CoA carboxylase beta chain | D [auth G], E [auth H], J [auth M], K [auth N] | 542 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 EC: 6.4.1.3 | 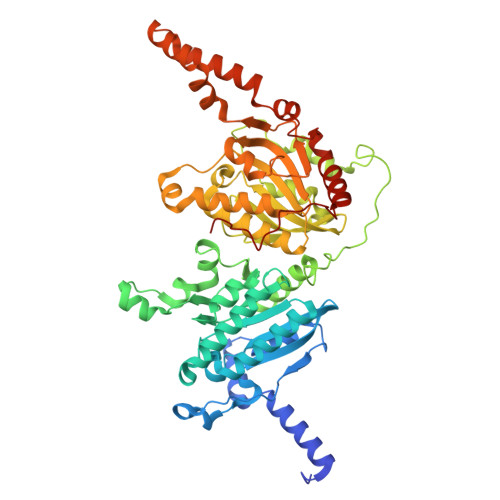 |
UniProt | |||||
Find proteins for A0QTE7 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0QTE7 Go to UniProtKB: A0QTE7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QTE7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophanate hydrolase subunit 2 | O [auth V], R [auth Y] | 294 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0QPL0 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0QPL0 Go to UniProtKB: A0QPL0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QPL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophanate hydrolase subunit 1 | P [auth W], S [auth Z] | 210 | Mycolicibacterium smegmatis MC2 155 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0QPL1 (Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)) Explore A0QPL1 Go to UniProtKB: A0QPL1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QPL1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1CZD (Subject of Investigation/LOI) Query on A1CZD | AA [auth F], HA [auth O] | S-{(3S,5S,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5,10,14-tetraoxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (11E,14E,16E)-icosa-11,14,16-trienethioate (non-preferred name) C41 H68 N7 O17 P3 S VXJQAOBDWILAOY-WVXUJWCUSA-N |  | ||
| 1VU (Subject of Investigation/LOI) Query on 1VU | BA [auth G], CA [auth H], FA [auth M], GA [auth N] | propionyl Coenzyme A C24 H40 N7 O17 P3 S QAQREVBBADEHPA-IEXPHMLFSA-N |  | ||
| ATP (Subject of Investigation/LOI) Query on ATP | KA [auth Q], OA [auth R] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| BTN (Subject of Investigation/LOI) Query on BTN | DA [auth I], EA [auth K], IA [auth Q], MA [auth X], NA [auth U] | BIOTIN C10 H16 N2 O3 S YBJHBAHKTGYVGT-ZKWXMUAHSA-N |  | ||
| BCT (Subject of Investigation/LOI) Query on BCT | JA [auth Q], QA [auth R] | BICARBONATE ION C H O3 BVKZGUZCCUSVTD-UHFFFAOYSA-M |  | ||
| MG (Subject of Investigation/LOI) Query on MG | LA [auth Q], PA [auth R] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19.2_4158 |
| RECONSTRUCTION | cryoSPARC | 5.0.0-privatebeta |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | PJT191893 |