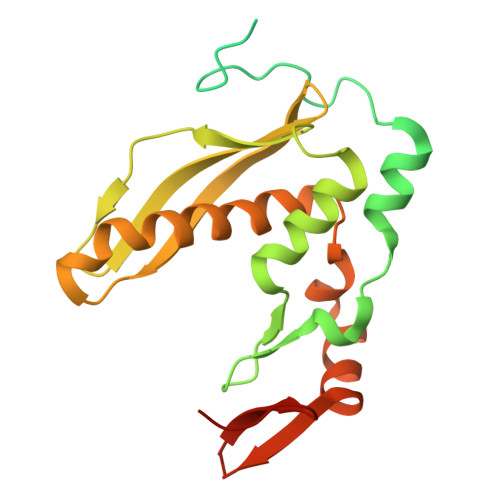
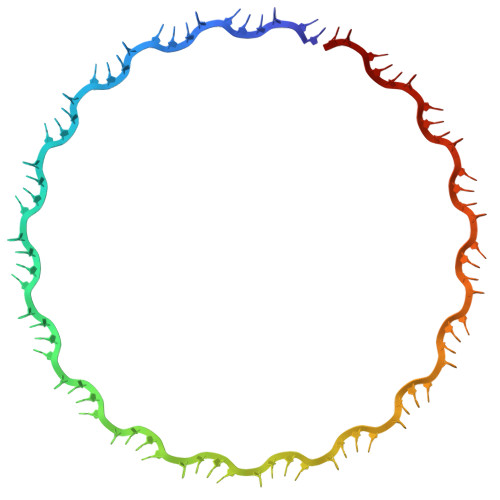
Mechanism of single-strand annealing from native mass spectrometry and cryo-EM structures of RAD52 homolog Mgm101.
Wheat, C.T., Qi, Z., Hussain, M., Zakharova, K., Wysocki, V.H., Bell, C.E.(2025) bioRxiv
- PubMed: 41280126
- DOI: https://doi.org/10.1101/2025.10.26.684629
- Primary Citation of Related Structures:
9YI6, 9YI7, 9YI8, 9YI9, 9YIA - PubMed Abstract:
RAD52, the primary single-strand annealing (SSA) protein in humans, forms undecameric rings that bind ssDNA within a narrow, positively-charged groove. Whether RAD52 anneals two complementary ssDNAs on the same ring in cis , or between two ring-ssDNA complexes in trans , is unknown. Here, we determined cryo-EM structures of Mgm101, a RAD52 homolog from yeast mitochondria, in complexes with ssDNA, a duplex intermediate of annealing, and B-form dsDNA product. In all states, Mgm101 forms a closed nonadecameric ring that binds the backbone of the first ssDNA at the base of the narrow groove. The second complementary strand binds directly on top of the first to form an extended, unwound, and circular duplex intermediate of annealing. The third complex captures apparent B-form DNA product bound to a novel β-hairpin motif located on top of the Mgm101 ring, above the primary DNA-binding groove. Mass photometry and native mass spectrometry confirm and further elucidate the complexes formed in solution. Altogether, our data reveal the full SSA pathway of Mgm101 and suggest it anneals two complementary ssDNAs on the same ring in cis . Structural conservation with RAD52 suggests it is likely to use a similar cis mechanism of annealing.
- Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: