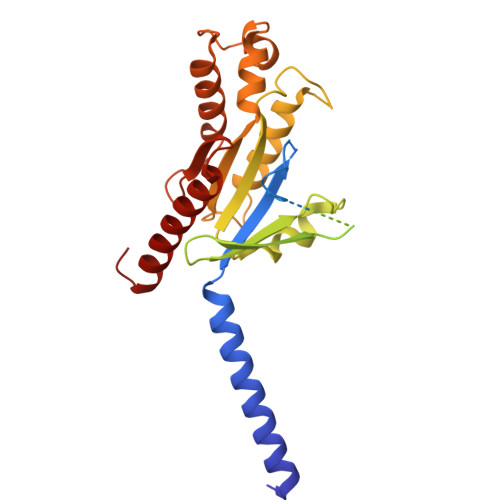
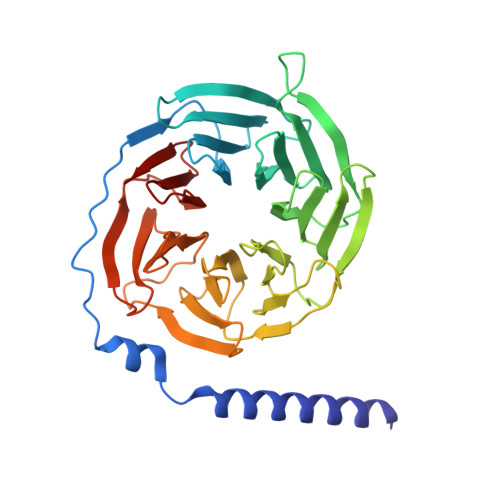
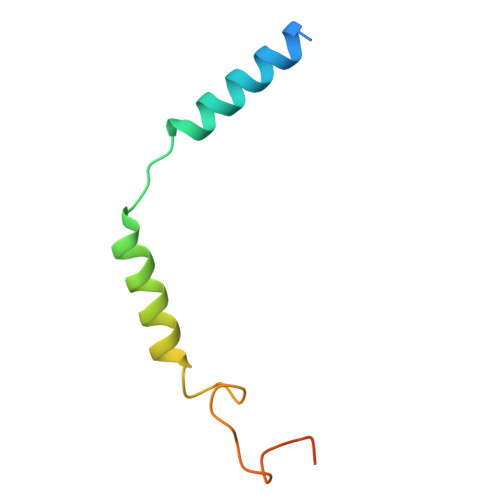
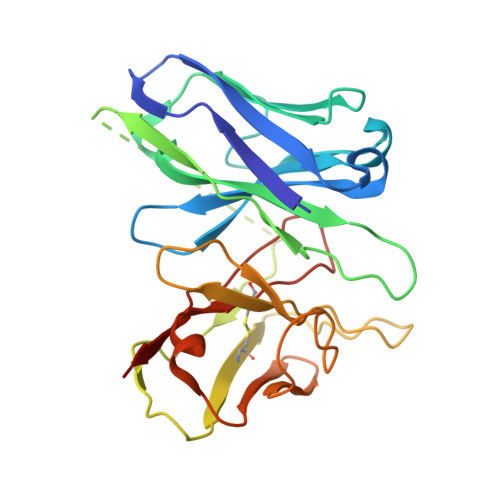
UDP-glucose and MRS2905 agonist-bound states of the purinergic P2Y 14 receptor.
Fay, J.F., Kousouros, J., Gao, Z.G., Pavan, M., Oliva, P., Pramanik, A., Meyer, C., Kurma, S.H., Xu, W., Krumm, B., Jacobson, K.A.(2025) Commun Biol 8: 1816-1816
- PubMed: 41272298
- DOI: https://doi.org/10.1038/s42003-025-09174-6
- Primary Citation of Related Structures:
9YDU, 9YDV - PubMed Abstract:
P2Y 14 receptors respond to uracil-diphosphate-hexose conjugates, yet how this receptor selectively recognizes both uracil-nucleotides and hexose moieties of diverse agonists remains unclear. Here we report the active agonist-bound G protein-bound states of the P2Y 14 G protein-coupled receptor (GPCR) bound to endogenous agonist uridine diphosphate glucose (UDP-glucose) and 2-thiouridine-5'-O-(α,β-methylene)diphosphate (MRS2905). The cryo-EM structures of the heterotrimeric complexes with 3.19 Å and 3.05 Å global resolution, respectively, with local refinements reaching 2.87 Å and 3.22 Å for the masked receptor region. Our structures reveal a pronounced extracellular facing electronegative vestibule connecting to a smaller nucleotide binding subpocket (~300Å 3 volume) that is shielded by extracellular loop 2 (ECL2). A glucose-binding subpocket is spatially delimited by residue V93; mutation to Trp selectively blocks UDP-glucose while permitting MRS2905 and antagonist binding. These findings provide atomic insights into uracil recognition, reveal how the receptor accommodates diverse flexible UDP-sugars, and promise to enable rational drug discovery of therapeutic P2Y 14 R modulators.
- Department of Biochemistry and Molecular Biology, University of Maryland Baltimore, Baltimore, MD, USA. jfay@som.umaryland.edu.
Organizational Affiliation: