Structure and Activity of a beta-Mannosidase Involved in Core N-Glycan Degradation.
Zea, G., Landry, J., Winchester, A., Sharma, S., Higgins, M.A.(2025) Proteins
- PubMed: 41376108
- DOI: https://doi.org/10.1002/prot.70106
- Primary Citation of Related Structures:
9Y4V - PubMed Abstract:
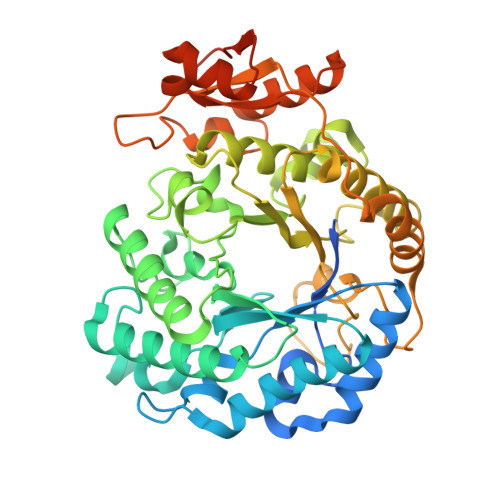
N-glycans are structurally complex carbohydrates commonly found on eukaryotic glycoproteins, where they play essential roles in protein folding, stability, and cellular signaling. Some bacteria have evolved specialized degradation pathways to access N-glycans as nutrient sources, terminating in enzymes that cleave the conserved core Manβ1-4GlcNAc disaccharide. Members of glycoside hydrolase family 5 subfamily 18 (GH5_18) have recently been identified to catalyze this reaction. Here, we report the biochemical and structural characterization of MoGH5_18, which is encoded within a gene cluster consisting of other genes likely involved in N-glycan degradation. Biochemical assays show that MoGH5_18 hydrolyzes Manβ1-4GlcNAc but not Manβ1-4Man, consistent with substrate specificity observed in other GH5_18s. We solved the crystal structure of MoGH5_18 to 1.92 Å resolution, revealing a canonical (β/α) 8 TIM-barrel fold, dimeric architecture, and a conserved active site architecture. These findings demonstrate that MoGH5_18, despite sequence divergence, retains the structural and functional hallmarks of GH5_18 enzymes and further illustrate the power of SSN-guided approaches to uncover conserved enzymatic mechanisms within diverse glycan degradation pathways.
- Department of Biological Sciences, The University of Alabama, Tuscaloosa, Alabama, USA.
Organizational Affiliation: