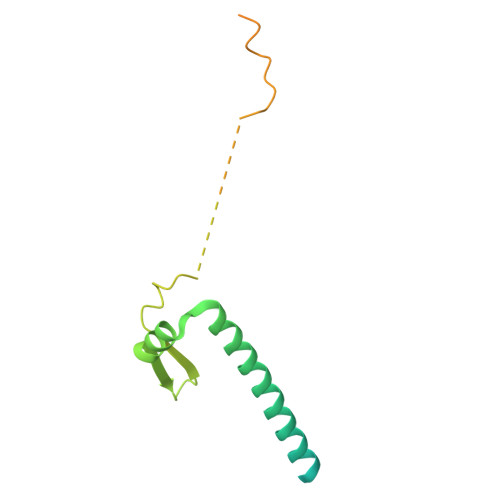
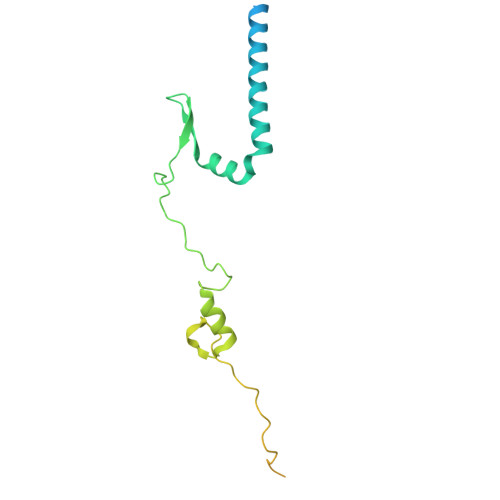
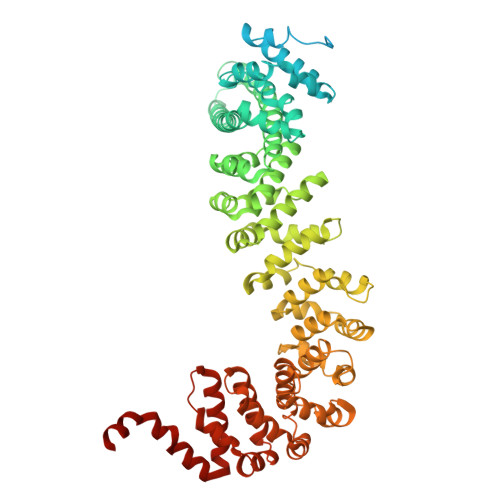
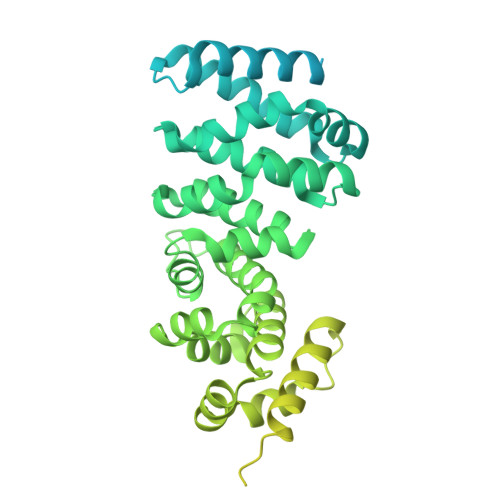
The hook-like adaptor and cargo-binding (HAC) domain in the kinesin-2 tail enables adaptor assembly and cargo recognition.
Jiang, X., Danev, R., Ichinose, S., Niu, B., Ohtsuki, S., Yanagisawa, H., Nagatoishi, S., Tsumoto, K., Hirokawa, N., Kikkawa, M.(2025) Sci Adv 11: eady5861-eady5861
- PubMed: 41134888
- DOI: https://doi.org/10.1126/sciadv.ady5861
- Primary Citation of Related Structures:
9W9H, 9W9I - PubMed Abstract:
Intracellular transport relies on motor proteins such as kinesins to deliver cargo along microtubules, yet how they recognize cargo remains unclear. Here, we present high-resolution cryo-electron microscopy structures of the heterotrimeric kinesin-2 complex (KIF3A/KIF3B/KAP3) bound to the cargo protein APC. Our findings reveal a previously uncharacterized KIF3 tail hook-like motif, termed the "HAC" domain, which mediates binding to both KAP3 adaptor and APC cargo. Within this domain, the KIF3A helical regions ensure cargo specificity, while a β-hairpin and KIF3B provide structural support. Biochemical and neuronal experiments confirm its functional importance. Notably, the HAC/KAP3 structure resembles hook-like architectures seen in kinesin-1 and dynein, suggesting a shared cargo recognition framework. These findings also shed light on kinesin-2 cargo specificity and offer a structural framework for understanding related neuronal transport mechanisms.
- Department of Cell Biology and Anatomy, Graduate School of Medicine, The University of Tokyo, Tokyo 113-0033, Japan.
Organizational Affiliation: