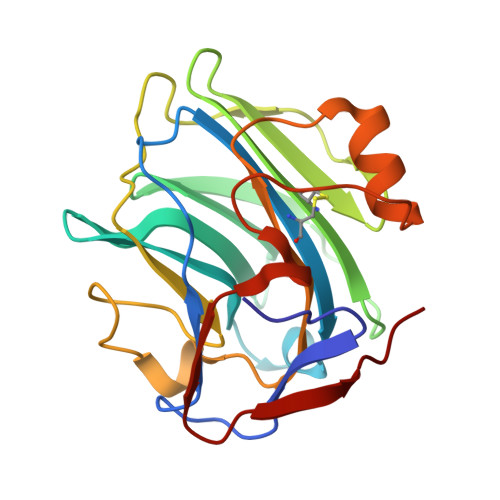
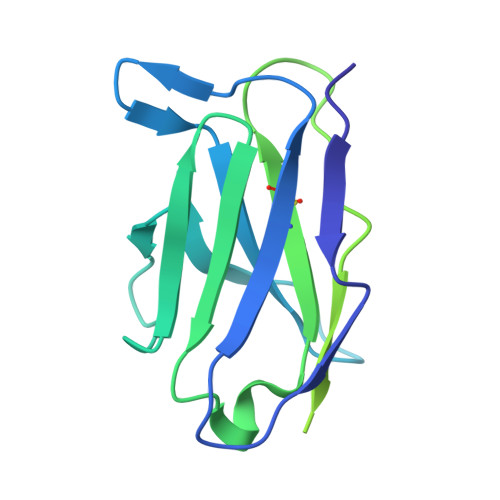
Cryo-EM structure of pentameric C-reactive protein in complex with monoclonal IgG antibodies.
Moiseenko, A.V., Kalikin, A.V., Orekhov, P.S., Byzova, N.A., Zherdev, A.V., Shaitan, K.V., Dzantiev, B.B., Sokolova, O.S.(2025) FEBS J
- PubMed: 41159871
- DOI: https://doi.org/10.1111/febs.70310
- Primary Citation of Related Structures:
9VCA - PubMed Abstract:
C-reactive protein (CRP) plays a central role in innate immunity and serves as a key biomarker of inflammation. Despite its clinical importance, the structural basis of CRP interactions with antibodies remains poorly characterized. Using cryo-electron microscopy (cryo-EM), we resolved the structure of immune complexes formed between pentameric CRP and monoclonal immunoglobulin G (IgG) antibodies at up to 2.4 Å resolution. The complexes display a barrel-shaped architecture, with two CRP pentamers bridged by three to five antibodies. We built an atomic model of the CRP-antibody interface, identifying a binding site on the A-face of CRP mediated exclusively by hydrogen bonds, without salt-bridge formation. These findings provide structural insights into CRP-IgG recognition and offer a basis for the rational design of improved antibodies.
- Faculty of Biology, Lomonosov Moscow State University, Moscow, Russia.
Organizational Affiliation: