Helical structure of KomBCMUT in complex with dITP and NAD
Li, Y., Zheng, Q., Li, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Xanthosine/inosine triphosphate pyrophosphatase | 181 | Archangium gephyra | Mutation(s): 0 Gene Names: AA314_06977 | 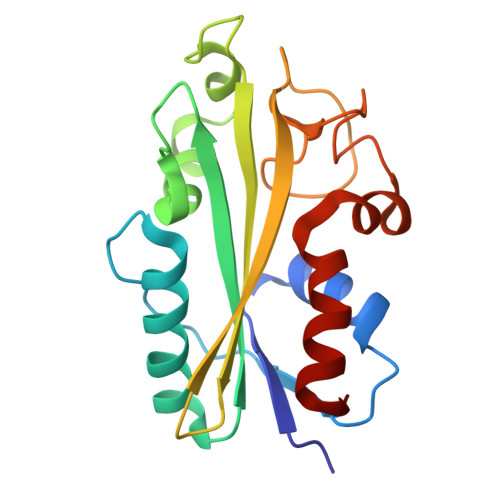 | |
UniProt | |||||
Find proteins for A0AAC8QDB0 (Archangium gephyra) Explore A0AAC8QDB0 Go to UniProtKB: A0AAC8QDB0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0AAC8QDB0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NAD-dependent protein deacetylase | 278 | Archangium gephyra | Mutation(s): 0 Gene Names: AA314_06978, ATI61_102412 | 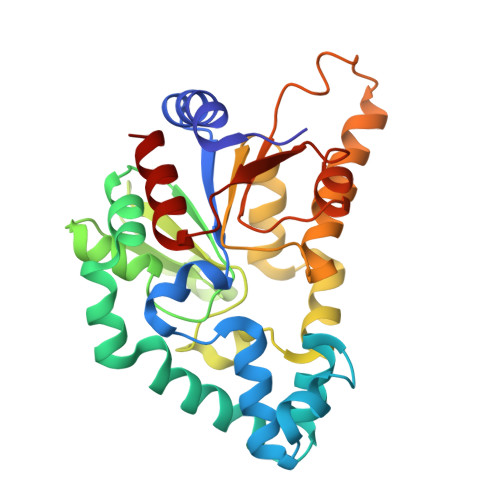 | |
UniProt | |||||
Find proteins for A0AAC8TGL7 (Archangium gephyra) Explore A0AAC8TGL7 Go to UniProtKB: A0AAC8TGL7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0AAC8TGL7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAD Query on NAD | GA [auth i] HA [auth j] IA [auth k] JA [auth l] KA [auth m] | NICOTINAMIDE-ADENINE-DINUCLEOTIDE C21 H27 N7 O14 P2 BAWFJGJZGIEFAR-NNYOXOHSSA-N |  | ||
| Y43 (Subject of Investigation/LOI) Query on Y43 | BA [auth T] DA [auth V] FA [auth X] Q [auth I] T [auth K] | 2'-deoxyinosine 5'-triphosphate C10 H15 N4 O13 P3 UFJPAQSLHAGEBL-RRKCRQDMSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | AA [auth T] CA [auth V] EA [auth X] R [auth I] S [auth K] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |