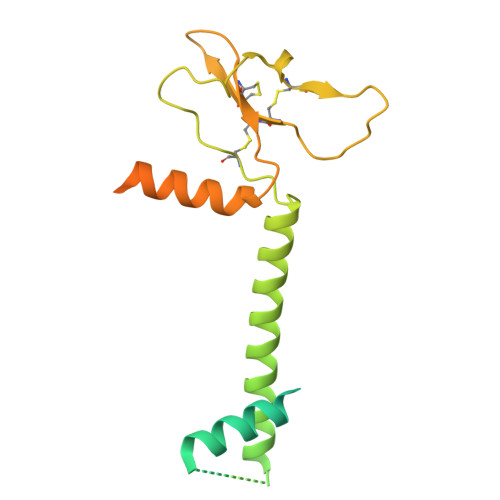
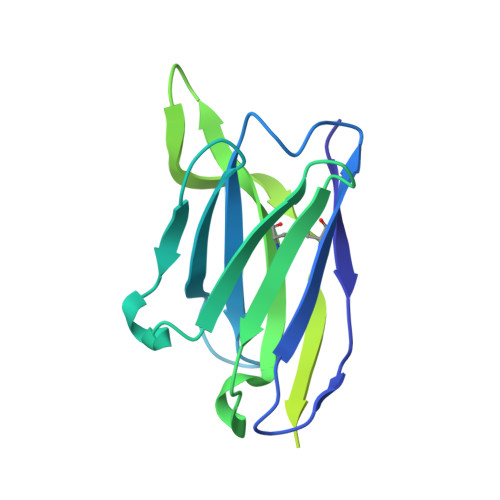
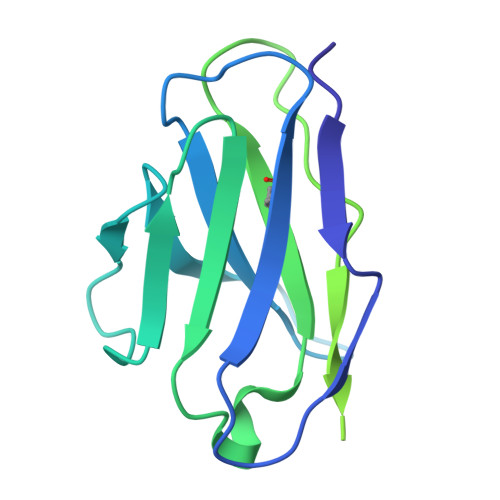
The Symmetric Structure of the Antigenic Loop in Type B HBV Surface Antigen.
Tao, W., He, X., Chen, L.(2025) J Mol Biology 437: 169483-169483
- PubMed: 41075972
- DOI: https://doi.org/10.1016/j.jmb.2025.169483
- Primary Citation of Related Structures:
9UBQ - PubMed Abstract:
Hepatitis B virus (HBV) is an enveloped virus with HBV surface antigen (HBsAg) as the only protein on its viral membrane. The extracellular antigenic loop (AGL) of HBsAg plays a crucial role in viral attachment to host cells, serves as the primary target for neutralizing antibodies (NAbs), and is subject to escape mutations. Previous studies have shown that the AGL exhibits two different structures (Type A and Type B) dictated by distinct disulfide bond linkage. However, due to the flexibility of some regions in previous structure, the complete model of AGL Type B and its symmetry remain elusive. Here, we present the cryo-EM structure of AGL Type B in complex with the Fab fragment of the NAb H020. The complete structure of AGL Type B reveals its two-fold symmetry and it can bind two Fab H020 fragments simultaneously. Further analysis elucidates the underlying mechanism of pan-serotype neutralizing capability of H020 and how escape mutations hinder its binding.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Peking. University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing 100871, China.
Organizational Affiliation: