High resolution crystal structures of duck RIG-I and chicken LGP2 bound to non-canonical duplexes mimicking the influenza virus vRNA promoter (panhandle).
Cusack, S., Ivanov, I., Uchikawa, E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA helicase | 692 | Anas platyrhynchos | Mutation(s): 0 Gene Names: RIG-I EC: 3.6.4.13 | 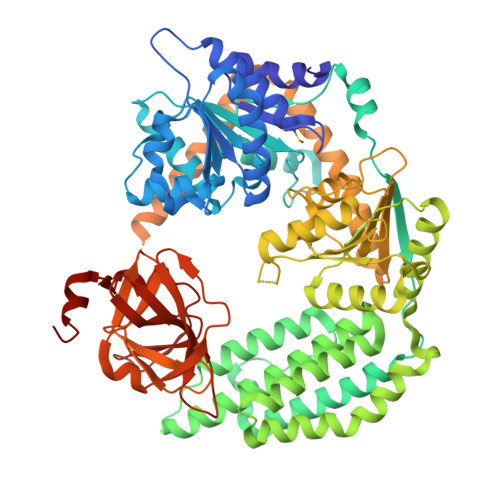 | |
UniProt | |||||
Find proteins for D3TI84 (Anas platyrhynchos) Explore D3TI84 Go to UniProtKB: D3TI84 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D3TI84 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 31-mer RNA hairpin with 5'pppG-C blunt end mimicking the influenza B virus vRNA promoter (panhandle) | B [auth R] | 30 | Influenza B virus | 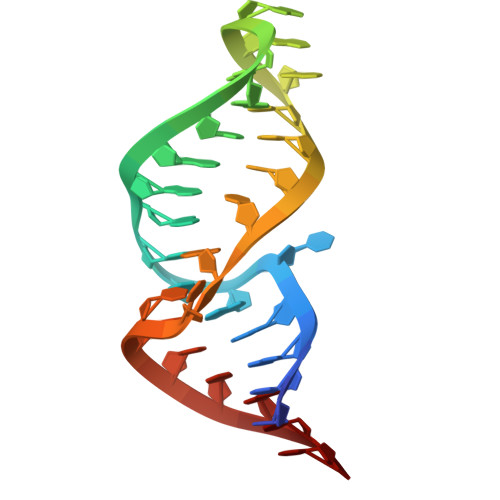 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP (Subject of Investigation/LOI) Query on GTP | J [auth R] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| GOL Query on GOL | I [auth R] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| K Query on K | F [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | D [auth A], G [auth A], H [auth R] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 87.641 | α = 90 |
| b = 100.072 | β = 90 |
| c = 192.287 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| STARANISO | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | France | -- |