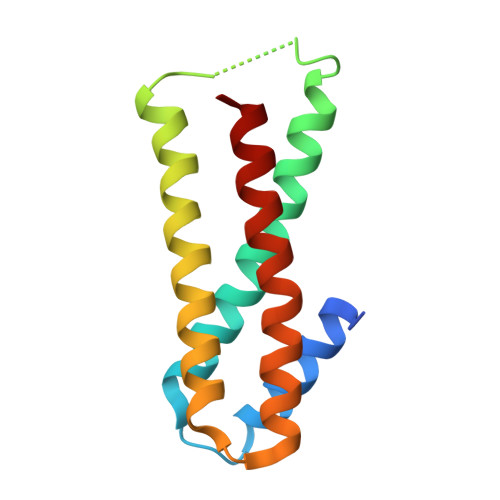
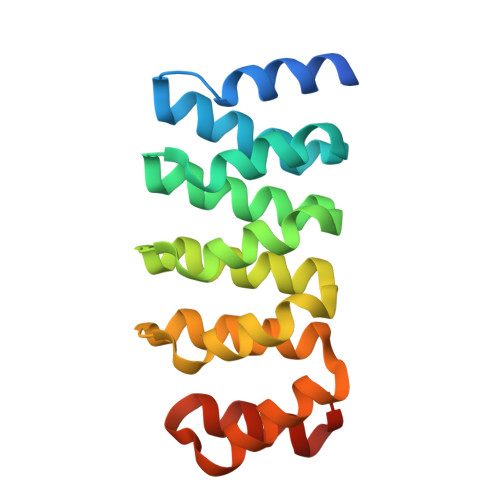Monomeric Structure of Influenza A Virus NEP/NS2 Obtained With an Artificial Protein Highlights Conformational Plasticity.
Stelfox, A.J., Bessonne, M., Bourhis, J.M., Erba, E.B., Albanese, P., Compte, D.P., Nevers, Q., Urvoas, A., Valerio-Lepiniec, M., Minard, P., Ruigrok, R.W.H., Crepin, T., Delmas, B., Ballandras-Colas, A.(2025) J Mol Biology 437: 169511-169511
- PubMed: 41175965
- DOI: https://doi.org/10.1016/j.jmb.2025.169511
- Primary Citation of Related Structures:
9T1M - PubMed Abstract:
The influenza virus nuclear export protein (NEP)/non-structural protein 2 (NS2) is a multifunctional protein, involved in viral ribonucleoprotein export from the nucleus, genome replication enhancement, and the adaption of avian influenza to mammals. Despite the growing attention on the importance of NEP in the influenza virus lifecycle and in interspecies transmission, the molecular details of how it performs its various roles are still not fully understood. For the purpose of assisting in structural characterization, a panel of artificial proteins (αReps) were selected against influenza virus A NEP H1N1 by phage display. When complexed with αRep E4, we were able to crystallize full-length NEP H1N1 , and characterize the NEP- αRep E4 interface using an integrative native MS-XL-MS strategy, revealing a folded and monomeric NEP conformation. The N- and C-termini of NEP H1N1 pack together, with the middle linker region resembling a hinge, contrasting a previous structure where NEP is dimeric and elongated. Together these structures demonstrate the plasticity of NEP, a trait which may potentially aid NEP in binding cellular and viral partners. Using isothermal titration calorimetry (ITC) we measured a nanomolar interaction between αRep E4 and NEP. Similarly, we found that αRep E4 also binds NEP from human-isolated avian H7N9 and bovine-isolated avian H5N1 influenza viruses. Owing to their high degree of conservation, αRep E4 likely has the capacity to interact with NEP from numerous influenza A virus strains. Indeed, this, combined with the nanomolar affinity measured between NEP/ αRep E4 could be explored further as a broad-range therapeutic strategy and/or a tool in cellulo to understand NEP function.
- Univ. Grenoble Alpes, CNRS, CEA, IBS, F-38000 Grenoble, France.
Organizational Affiliation: