Doxorubicin Recognition and Transport by the MATE Multidrug Transporter NorM From Vibrio cholerae.
Hsieh, P.Y., Romane, K., Kowal, J., Locher, K.P., van Veen, H.W.(2025) J Mol Biology 438: 169549-169549
- PubMed: 41260293
- DOI: https://doi.org/10.1016/j.jmb.2025.169549
- Primary Citation of Related Structures:
9RSJ - PubMed Abstract:
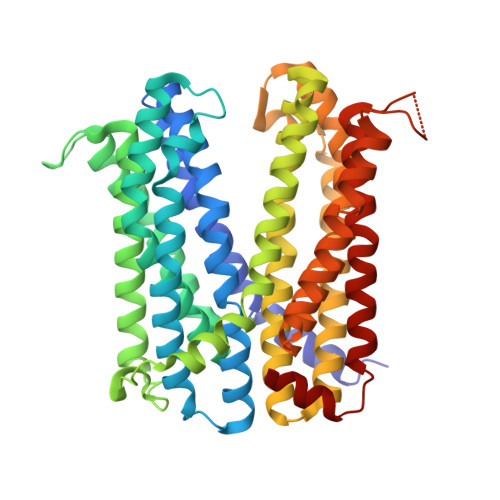
Multidrug and toxic compound extrusion (MATE) transport proteins contribute to multidrug resistance in human pathogens by extruding various cytotoxic compounds from the cellular interior. Despite their importance across all domains of life, the specificities and mechanisms of substrate transport of these proteins remain poorly understood due to limited structural and functional information. Here, we determined the cryo-electron microscopy structure of NorM from Vibrio cholerae (NorM-VC) in complex with the anthracycline antibiotic doxorubicin, using the NabFab approach. The structure reveals that the doxorubicin-binding pocket is located halfway through the membrane, within the C-lobe of the protein. Functional studies targeting the doxorubicin-interacting residues validated the binding pocket and enabled detailed analysis of the doxorubicin transport reaction. Our findings indicate doxorubicin binding within a multisite binding chamber engaged in a general transport mechanism for a variety of substrates.
- Department of Pharmacology, University of Cambridge, Cambridge CB2 1PD, UK.
Organizational Affiliation: