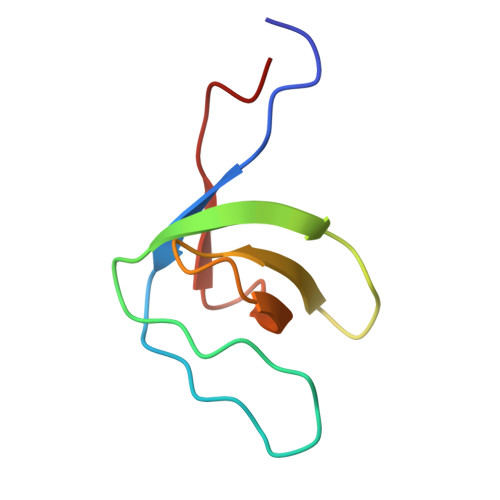
Intrinsically disordered enteropathogenic E. coli EspF exploits motif mimicry in high-affinity binding to neural Wiskott-Aldrich syndrome protein and sorting nexin 9.
Tossavainen, H., Karjalainen, M., Antenucci, L., Hellman, M., Permi, P.(2025) Int J Biol Macromol 330: 148227-148227
- PubMed: 41075884
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.148227
- Primary Citation of Related Structures:
9R3Y, 9R4V, 9R4W - PubMed Abstract:
EspF is an enteropathogenic Escherichia coli (EPEC) effector protein that interferes with intestinal epithelial cell signaling by binding to the Src homology 3 (SH3) domain of sorting nexin 9 (SNX9) and the GTPase-binding domain (GBD) of neural Wiskott-Aldrich syndrome protein (N-WASP) with its C-terminal proline-rich repeats. To understand the molecular basis of these interactions, we characterized the structure, dynamics, and binding thermodynamics of EspF and its target protein domain complexes. We also elaborated on our previous study on EspF U , a homologous effector in enterohemorrhagic E. coli (EHEC), and compared the two effectors. We show that EspF is intrinsically disordered but that NMR chemical shifts expose the pre-structured polyproline II (PPII) helical SH3- and helical GBD-binding motifs. These motifs mimic their cellular counterparts but are fine-tuned to prevail in competitive binding. Factors behind EspF's higher affinity for GBD relative to the cellular ligand are key residue mutations and a C-terminally elongated polar interaction interface. The latter compensates for the lack of an "extended arm", the critical substitution promoting high affinity for GBD in EspF U . With this advantage, EspF outcompetes the autoinhibitory N-WASP C-helix and stimulates actin polymerization. EspF binds SNX9 SH3 with an extended binding interface, residues N-terminal to the RxAPxxP core motif being essential to strong binding. We define the SNX9 SH3-binding epitope as ϕxPxRxAPxxP and propose to re-delineate the EPEC EspF repeat boundaries accordingly. Furthermore, a characteristic 13 C secondary chemical shift pattern is recognized as a fingerprint of polyproline II (PPII) helical conformation in the SH3 binding epitope.
- Department of Biological and Environmental Science, Nanoscience Center, University of Jyväskylä, P.O. Box 35, FI-40014, Jyväskylä, Finland.
Organizational Affiliation: