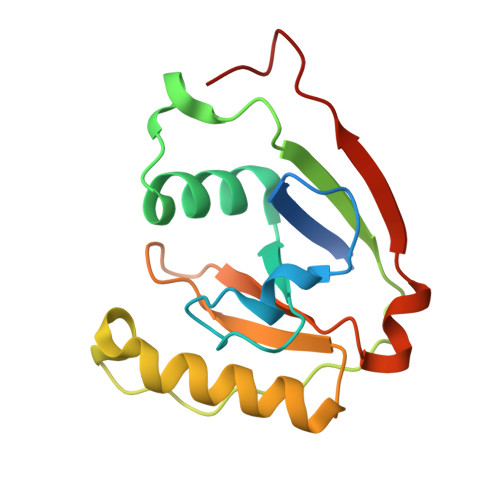
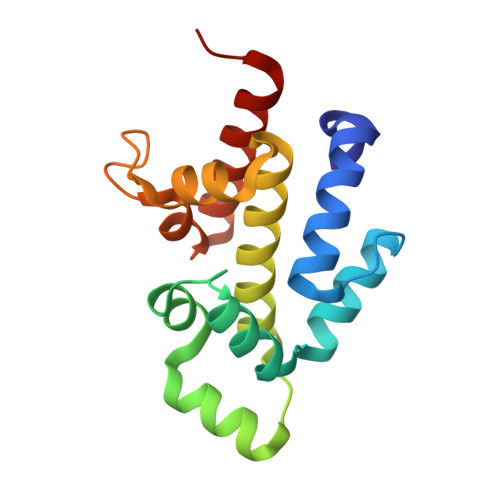
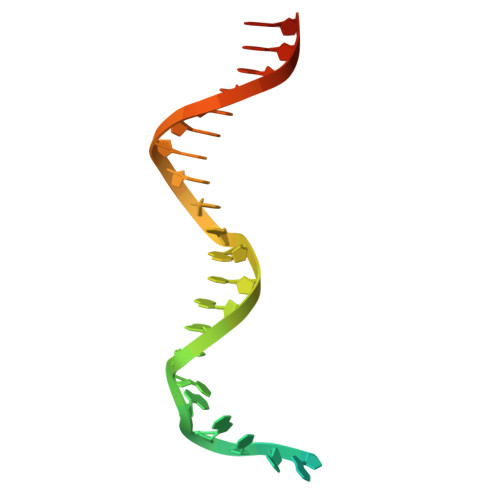
Structural basis for higher-order DNA binding by a bacterial transcriptional regulator.
Henriksen, F.O.G., Van, L.B., Brodersen, D.E., Skjerning, R.B.(2025) PLoS Genet 21: e1011749-e1011749
- PubMed: 40577318
- DOI: https://doi.org/10.1371/journal.pgen.1011749
- Primary Citation of Related Structures:
9R35 - PubMed Abstract:
Transcriptional regulation by binding of transcription factors to palindromic sequences in promoter regions is a fundamental process in bacteria. Some transcription factors have multiple dimeric DNA-binding domains, in principle enabling interaction with higher-order DNA structures; however, mechanistic and structural insights into this phenomenon remain limited. The Pseudomonas putida toxin-antitoxin (TA) system Xre-RES has an unusual 4:2 stoichiometry including two potential DNA-binding sites, compatible with a complex mechanism of transcriptional autoregulation. Here, we show that the Xre-RES complex interacts specifically with a palindromic DNA repeat in the promoter in a 1:1 molar ratio, leading to transcriptional repression. We determine the 2.7 Å crystal structure of the protein-DNA complex, revealing an unexpected asymmetry in the interaction and suggesting the presence of a secondary binding site, which is supported by structural prediction of the binding to the intact promoter region. Additionally, we show that the antitoxin can be partially dislodged from the Xre-RES complex, resulting in Xre monomers and a 2:2 Xre-RES complex, neither of which repress transcription. These findings highlight a dynamic, concentration-dependent model of transcriptional autoregulation, in which the Xre-RES complex transitions between a non-binding (2:2) and a DNA-binding (4:2) form.
- Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: