Structure of vaccinia virus A26 (residues 1-397) in complex with Fab 8M2110
Guardado-Calvo, P., Battini, L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelop protein OPG153 | 457 | Vaccinia virus Western Reserve | Mutation(s): 0 Gene Names: OPG153, VACWR149, A26L | 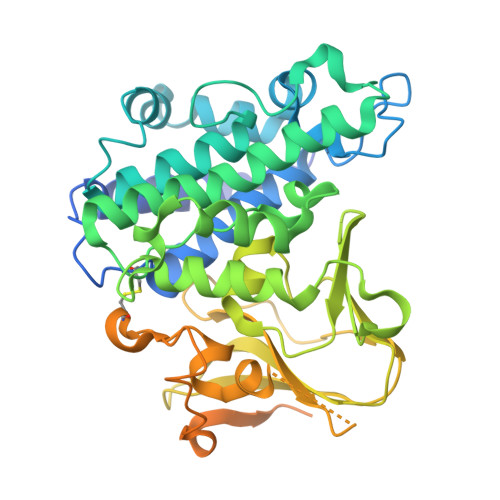 | |
UniProt | |||||
Find proteins for P24758 (Vaccinia virus (strain Western Reserve)) Explore P24758 Go to UniProtKB: P24758 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P24758 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heavy chain of Fab 8M2110 | 225 | Homo sapiens | Mutation(s): 0 | 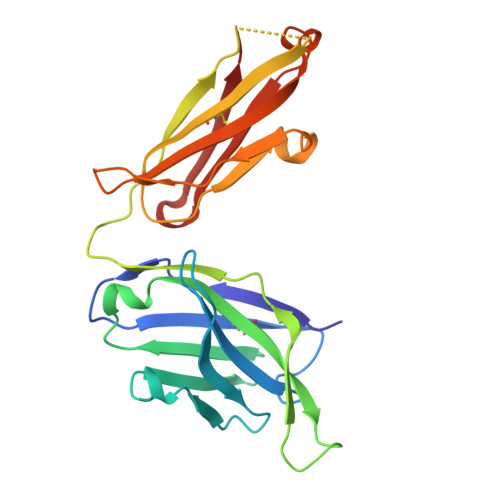 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Light chain of Fab 8M2110 | 214 | Homo sapiens | Mutation(s): 0 | 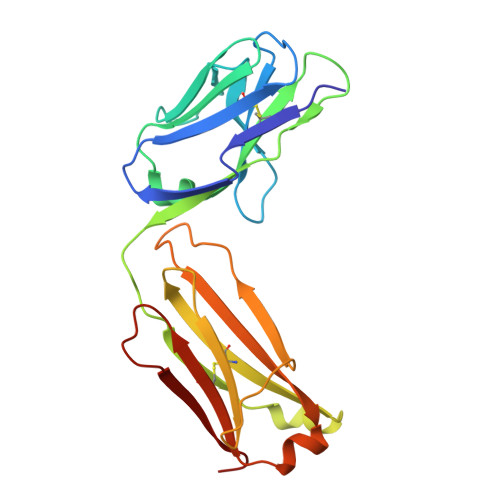 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anti-Fab VHH | 123 | Lama glama | Mutation(s): 0 | 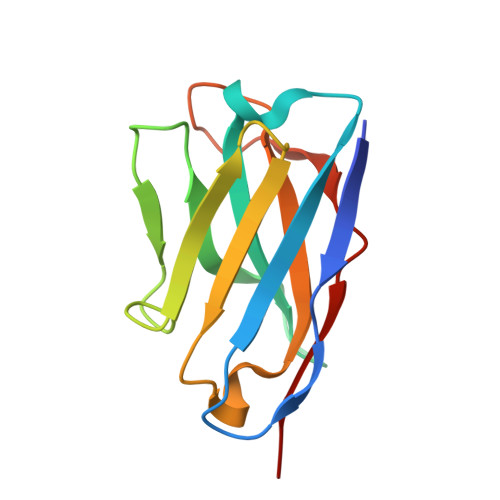 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 119.405 | α = 90 |
| b = 119.405 | β = 90 |
| c = 632.812 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Agence Nationale de la Recherche (ANR) | France | ANR-22-CE11-0003 |