The mechanistic basis of cargo selection during Golgi maturation.
Taylor, R.J., Zubkov, N., Ciazynska, K.A., Kaufman, J.G.G., Tagiltsev, G., Owen, D.J., Briggs, J.A.G., Munro, S.(2025) Sci Adv 11: eaea0016-eaea0016
- PubMed: 41042862
- DOI: https://doi.org/10.1126/sciadv.aea0016
- Primary Citation of Related Structures:
9QPQ - PubMed Abstract:
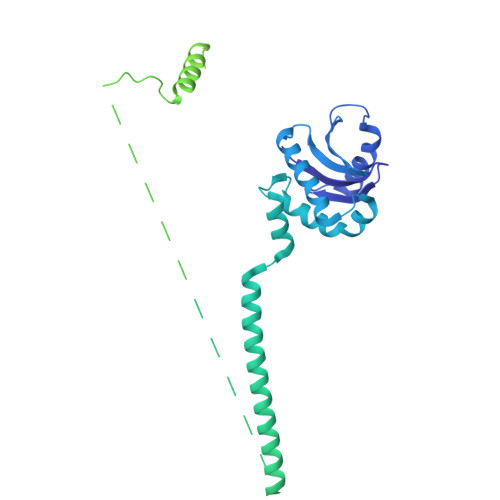
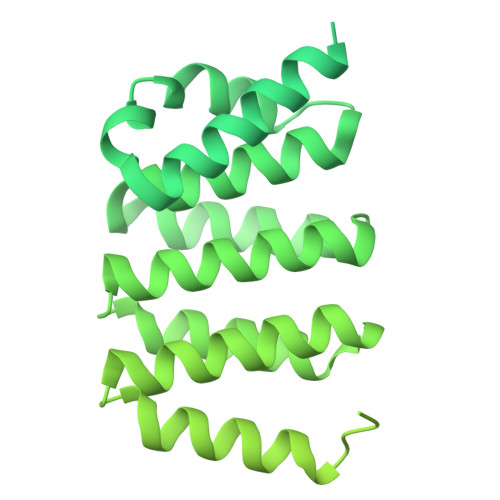
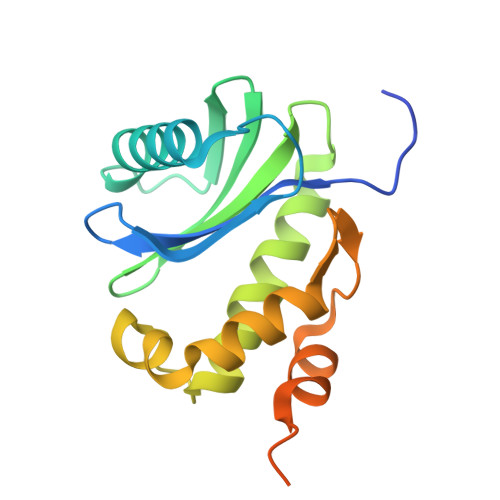
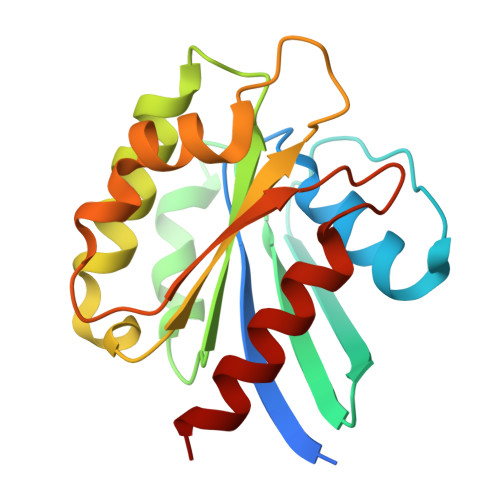
The multiple cisternae of the Golgi apparatus contain resident membrane proteins crucial for lipid and protein glycosylation. How Golgi residents remain in their designated compartments despite a constant flow of secretory cargo is incompletely understood. Here, we determine the structure of the COPI vesicle coat containing GOLPH3, an adaptor protein that binds the cytosolic tails of many Golgi residents. Analysis of this structure, together with structure-guided mutagenesis and functional assays, reveals how GOLPH3 uses coincidence detection of COPI and lipids to engage Golgi residents preferentially at late cisternae. Our findings rationalize the logic of cisternal maturation and explain how COPI can engage different types of substrates in different Golgi cisternae to retrieve some proteins back to the ER while retaining others within the Golgi apparatus.
- Department of Cell and Virus Structure, Max Planck Institute of Biochemistry, Martinsried, 82512, Germany.
Organizational Affiliation: