DNA polymerase with inhibitor
Lamers, M.H., Urem, M., Smits, W.K.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase III PolC-type | 1,474 | Enterococcus faecium | Mutation(s): 2 Gene Names: polC, CI258_000840 EC: 2.7.7.7 | 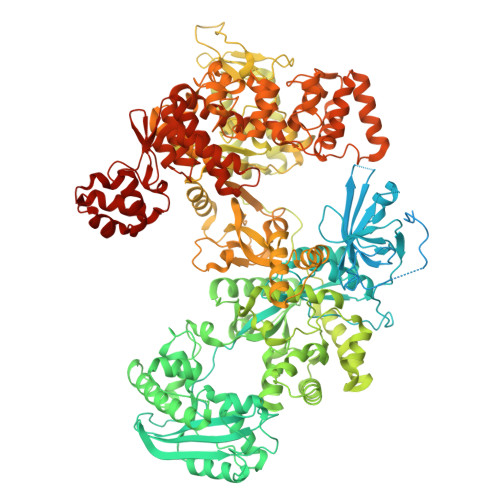 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*TP*AP*A)-3') | B [auth E] | 3 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*TP*AP*GP*GP*TP*GP*CP*GP*CP*AP*T)-3') | C [auth P] | 33 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*AP*CP*CP*TP*A)-3') | D [auth T] | 42 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1I88 Query on A1I88 | E [auth A] | 5-[[2,2-bis(fluoranyl)-1,3-benzodioxol-5-yl]methylamino]-3-fluoranyl-1-(2-hydroxyethyl)-6~{H}-pyrazolo[4,3-d]pyrimidin-7-one C15 H12 F3 N5 O4 KHEIVJSMPVGALA-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | F [auth A], G [auth A], H [auth A], I [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | J [auth A], K [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.18.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | Netherlands | POLSTOP2 |