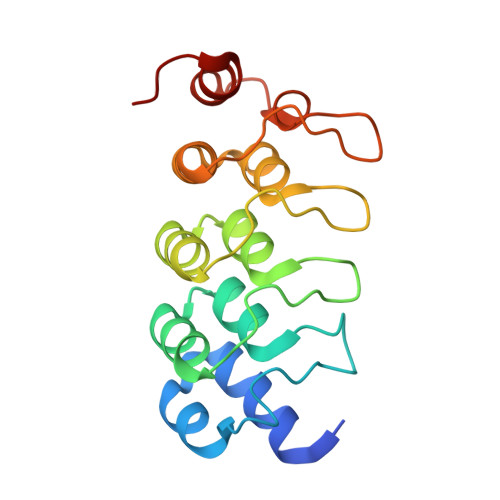
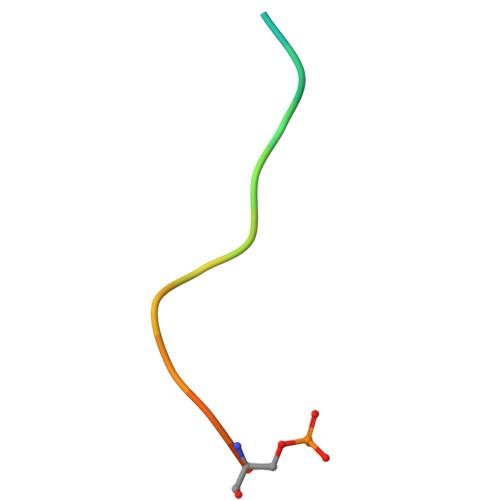
Phosphorylation as a candidate regulatory mechanism for effector recruitment to tankyrase.
Broadway, B.J., Pollock, K., Cronin, N., Rottapel, R., Sicheri, F., Guettler, S.(2025) R Soc Open Sci 12: 250824-250824
- PubMed: 41103766
- DOI: https://doi.org/10.1098/rsos.250824
- Primary Citation of Related Structures:
9QJZ - PubMed Abstract:
The ADP-ribosyltransferase tankyrase (with two paralogues, TNKS and TNKS2) plays pivotal roles in diverse cellular processes that encompass signal transduction, including Wnt/β-catenin, Hippo and toll-like receptor (TLR) signalling, mitotic spindle assembly, glucose homeostasis and telomere maintenance, among many other functions. Tankyrase recruits its effectors (substrates and binders) via a degenerate tankyrase-binding motif (TBM) and exerts its activities by subsequent substrate ADP-ribosylation and/or scaffolding. Variants of the TBM, found in diverse proteins, engage the ankyrin repeat cluster (ARC) domains of tankyrase. Yet, whether effector recruitment to tankyrase can be regulated has remained unknown. In this study, we propose that phosphorylation at position eight of the TBM enhances the affinity of effectors for the ARC domains of tankyrase. Using isolated TBM peptides, we demonstrate that phosphorylation of serine, but not tyrosine, strengthens ARC binding by up to an order of magnitude. Interrogation of proteome-wide phosphorylation data reveals that phosphorylation at position eight in the TBM is enriched in proteins that support centrosome function/localization. Our findings suggest that TBM phosphorylation may serve as an effector-specific mechanism for tankyrase recruitment/retention, providing an additional layer of regulation to control tankyrase.
- Division of Structural Biology, The Institute of Cancer Research, London, UK.
Organizational Affiliation: