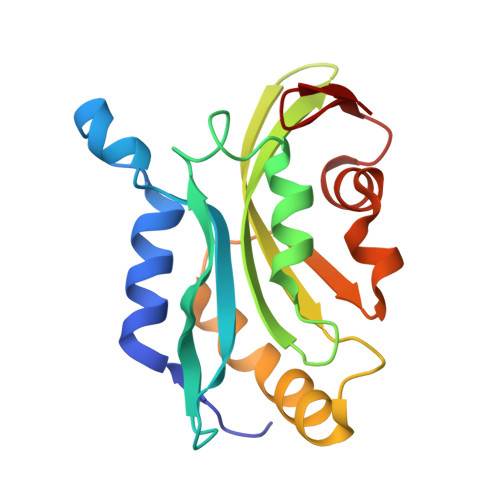
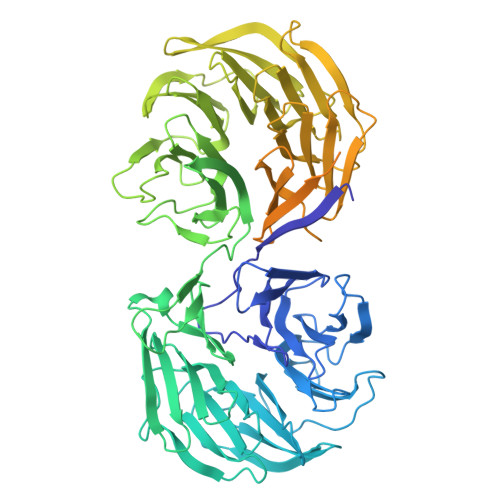
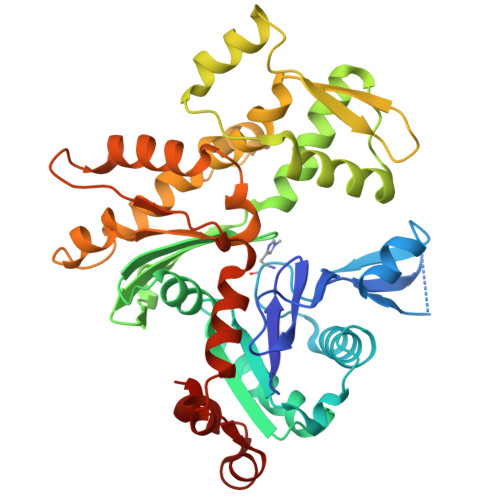
Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Oosterheert, W., Boiero Sanders, M., Hofnagel, O., Bieling, P., Raunser, S.(2025) Cell 188: 6845
- PubMed: 41075793
- DOI: https://doi.org/10.1016/j.cell.2025.09.016
- Primary Citation of Related Structures:
9QEW, 9QEY, 9QF2, 9QFB, 9QFD, 9QFE, 9QFG, 9QFJ, 9QFK, 9QFO, 9QFQ, 9QFW - PubMed Abstract:
Rapid remodeling of actin filament (F-actin) networks is essential for the movement and morphogenesis of eukaryotic cells. The conserved actin-binding proteins coronin, cofilin, and actin-interacting protein 1 (AIP1) act in synergy to promote rapid F-actin network disassembly, but the underlying mechanisms have remained elusive. Here, using cryo-electron microscopy (cryo-EM), we uncover the concerted molecular actions of coronin, cofilin, and AIP1 that lead to actin filament aging and severing. We find that the cooperative binding of coronin allosterically promotes inorganic phosphate release from F-actin and induces filament undertwisting, thereby priming the filament for cofilin binding. Cofilin then displaces coronin from the filament via a strand-restricted cooperative binding mechanism. The resulting cofilactin serves as a high-affinity platform for AIP1, which induces severing by acting as a clamp that disrupts inter-subunit filament contacts. In this "molecular squeezing" mechanism, AIP1 and not cofilin is responsible for filament severing. Our work redefines the role of key disassembly factors in actin dynamics.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, 44227 Dortmund, Germany. Electronic address: w.oosterheert@nki.nl.
Organizational Affiliation: