Structure of the Jeilongvirus Receptor Binding Receptor Protein Provides a Molecular Basis for Elongation of the Paramyxoviral C-terminus
Stelfox, A.J., Javorsky, A., Stass, R., Sutton, G., El Omari, K., Bowden, T.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Attachment glycoprotein | A [auth B], B [auth A] | 541 | Jeilongvirus beilongi | Mutation(s): 0 | 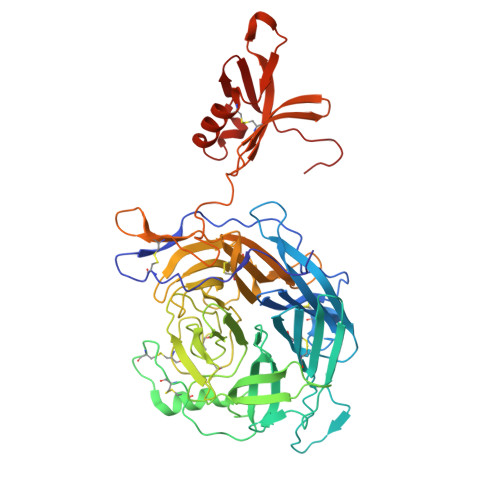 |
UniProt | |||||
Find proteins for Q287X2 (Beilong virus) Explore Q287X2 Go to UniProtKB: Q287X2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q287X2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | D [auth B], E [auth B], F [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| BMA Query on BMA | G [auth B] | beta-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-RWOPYEJCSA-N |  | ||
| MAN Query on MAN | H [auth B] | alpha-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-PQMKYFCFSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 87.98 | α = 90 |
| b = 162.38 | β = 90 |
| c = 226.07 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/L009528/1 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/S007555/1 |
| Engineering and Physical Sciences Research Council | United Kingdom | EP/K503113/1 |
| Engineering and Physical Sciences Research Council | United Kingdom | EP/L505031/1 |
| Engineering and Physical Sciences Research Council | United Kingdom | EP/M50659X/1 |
| Engineering and Physical Sciences Research Council | United Kingdom | EP/M508111/1 |
| Wellcome Trust | United Kingdom | 203141/Z/16Z |