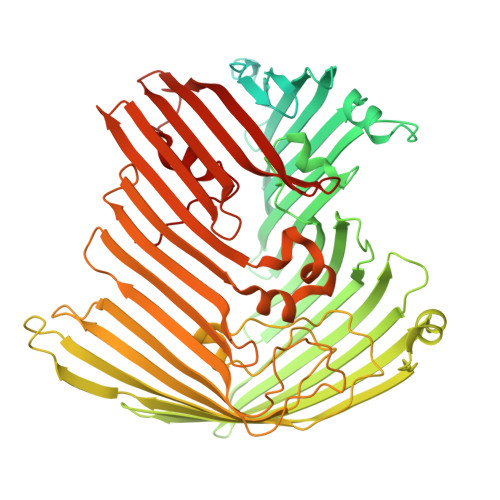
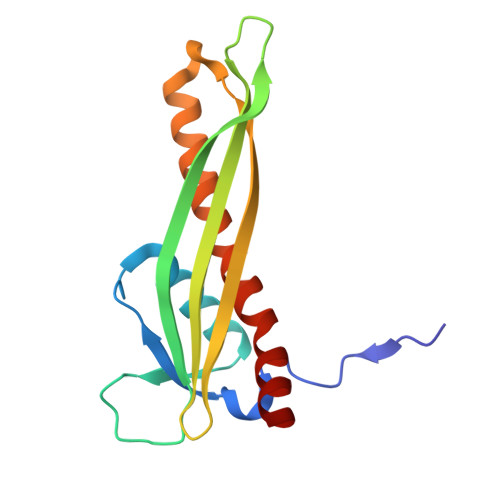
High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
Allyjaun, S., Dunbar, E., Hardwick, S.W., Newell, S., Holding, F., Rowland, C.E., St Denis, M.A., Pellegrino, S., Arruda Bezerra, G., Bournakas, N., Chirgadze, D.Y., Cooper, L., Paris, G., Lewis, N., Brown, P., Skynner, M.J., Dawson, M.J., Beswick, P., Hubbard, J., van den Berg, B., Newman, H.(2025) J Med Chem 68: 21144-21155
- PubMed: 41048016
- DOI: https://doi.org/10.1021/acs.jmedchem.5c00307
- Primary Citation of Related Structures:
9I92, 9I93, 9I94, 9I95, 9I96, 9I97, 9I98, 9Q8N - PubMed Abstract:
The lipopolysaccharide (LPS) transport (Lpt) system in Gram-negative bacteria maintains the integrity of the asymmetric bacterial outer membrane (OM). LPS biogenesis systems are essential in most Gram-negative bacteria, with LptDE responsible for the delivery of LPS to the outer leaflet of the OM. As an externally accessible, essential protein, LptDE offers a promising target for inhibitor development without the need for cellular penetration. However, there are no direct inhibitors of E. coli LptDE, and drug discovery is made challenging since it is a membrane target without a conventional active site. Here, the bicycle phage display platform was used in combination with cryogenic-electron microscopy (cryo-EM) and surface plasmon resonance to identify and map bicyclic peptide binders to Shigella flexneri LptDE (SfLptDE). Four distinct epitopes with unique bicycle molecule binding motifs were identified across the SfLptD β-barrel. This method represents a streamlined workflow for the identification and prioritization of hit molecules against LptDE.
- Biosciences Institute, Faculty of Medical Sciences, Newcastle University, Newcastle upon Tyne NE2 4HH, U.K.
Organizational Affiliation: