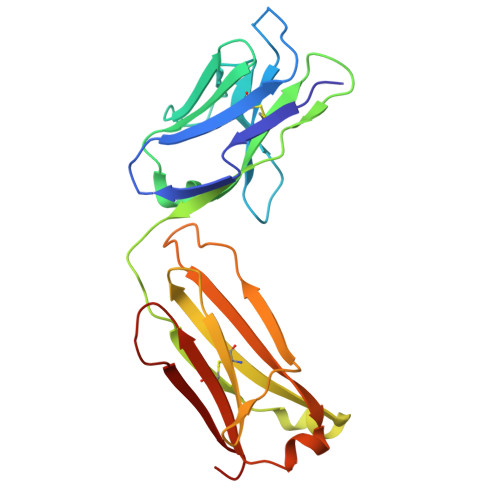
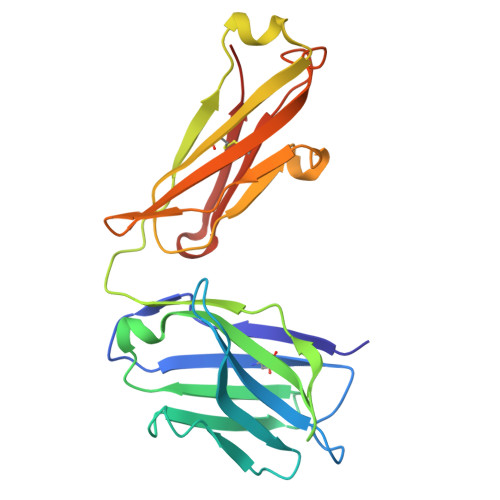
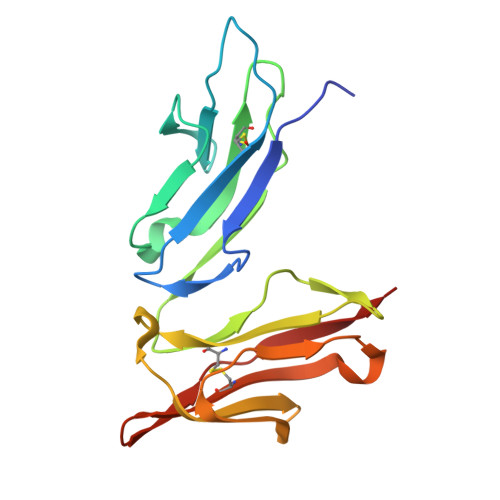
The impact of N-glycan conformational entropy on the binding affinity of Fc gamma receptor IIIa/CD16a.
Kremer, P.G., Tolbert, W.D., Gazaway, E., Hernandez, B.G., Korzeniowski, M.K., Dyba, Z.A., Grelsson, T., Grant, O.C., Lanzilotta, W.N., Pazgier, M., Woods, R.J., Barb, A.W.(2025) Structure
- PubMed: 41421343
- DOI: https://doi.org/10.1016/j.str.2025.11.015
- Primary Citation of Related Structures:
7URU, 9PRU - PubMed Abstract:
The affinity of Fc γ receptor IIIa (FcγRIIIa) binding to immunoglobulin G1 (IgG1) correlates with patient responses for antibody-based therapeutics. Among multiple factors affecting affinity, a mechanism defining how the composition of the FcγRIIIa N162 glycan regulates affinity remains undefined. Here, we evaluate the binding modes of two competitive FcγRIIIa ligands. IgG1 Fc binding is sensitive to N162 glycan composition, unlike the antigen-binding fragment (Fab) of the FcγRIII-specific antibody 3G8. Both ligands bound to overlapping surfaces, utilizing different angles of attack such that the IgG1 Fc but not 3G8 Fab limited the space available to the FcγRIII N162 N-glycan. FcγRIII binding to IgG1 Fc generated a 2.1 kcal/mol penalty from a loss of N162 glycan conformational entropy, greater than the 0.3 kcal/mol penalty for 3G8 and consistent with binding measurements. Thus, the conformational entropy of the FcγRIIIa N162-glycan is the predominant force modulating differential binding affinity compared to 3G8 Fab binding for endogenous FcγRIIIa glycoforms.
- Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, USA.
Organizational Affiliation: