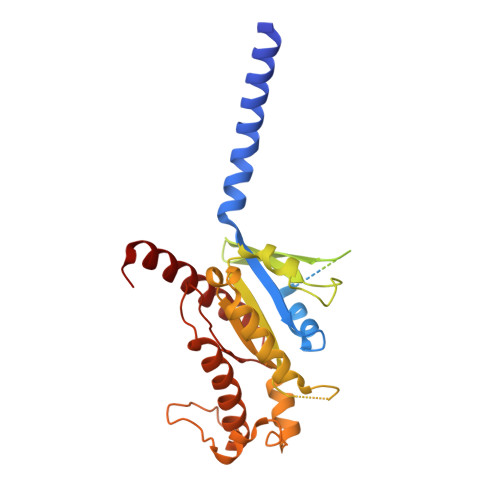
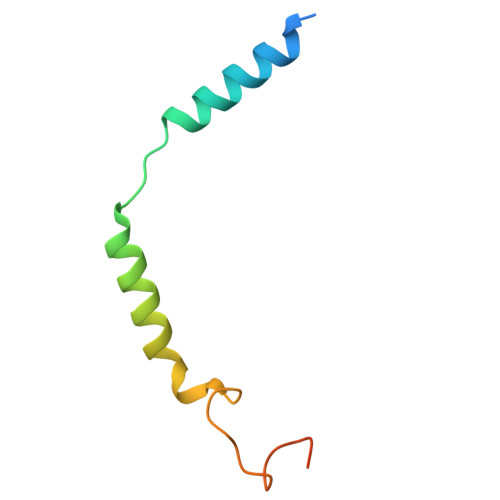
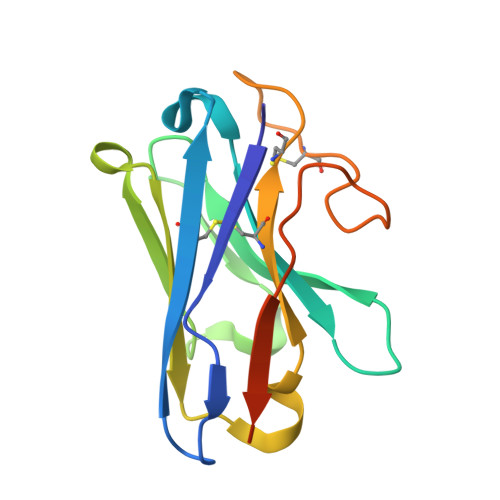
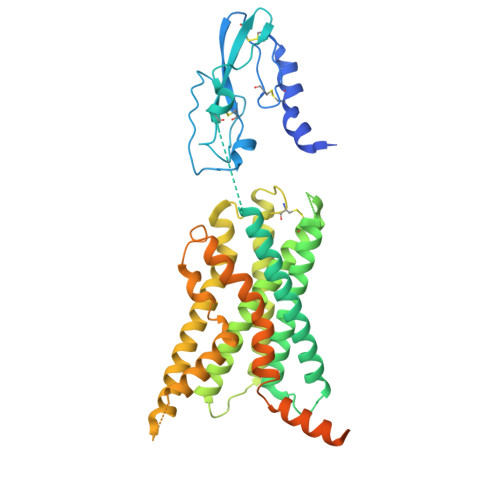
Structural basis of modified ligand selectivity from N-terminal PAC1R alternative splicing.
Lu, J.J., Deganutti, G., Li, M., Humphrys, L.J., Li, Y., Nettleton, T.J., Venugopal, H., Julita, V., Christopoulos, G., Reynolds, C.A., Sexton, P.M., Wootten, D., Zhao, P., Piper, S.J.(2025) Proc Natl Acad Sci U S A 122: e2521157122-e2521157122
- PubMed: 41264251
- DOI: https://doi.org/10.1073/pnas.2521157122
- Primary Citation of Related Structures:
9P92, 9P93, 9P94 - PubMed Abstract:
The pituitary adenylate cyclase-activating polypeptide (PACAP) 1 receptor (PAC1R) is a class B1 G protein-coupled receptor activated by the endogenous peptide agonists PACAP and vasoactive intestinal peptide (VIP). Alternate splicing within the receptor extracellular domain (ECD) generates the PAC1R short variant (PAC1sR) that has selectively enhanced VIP function compared to the full-length, PAC1R null variant (PAC1nR). However, to date, a comprehensive pharmacological assessment of the downstream signaling outcomes of PAC1sR activation compared to PAC1nR has not been performed, and little information is available to mechanistically understand how ECD splicing may alter ligand engagement. Here, we demonstrated that VIP, but not PACAP, has globally enhanced activity across a broad range of functional endpoints at PAC1sR compared to PAC1nR. Cryo-EM structures of VIP-bound, stimulatory G protein (G s )-coupled PAC1sR and PAC1nR, supported by molecular dynamics (MD) simulations, demonstrate transient engagement of the null loop in PAC1nR, which is absent in PAC1sR, with residues in extracellular loop 2 (ECL2) and the N-terminal helix of the ECD. These interactions result in differential engagement of VIP with these domains and the top of TM2/ECL1 with PAC1sR and PAC1nR. Moreover, MD simulations predicted differential interactions of the G s protein with the two PAC1R variants when bound by VIP that correlate with a greater allosteric influence of the G s protein on VIP affinity at the PAC1sR, relative to PAC1nR. Our study provides insights into the structural basis and functional consequences of PAC1R ECD splicing, increasing understanding of PAC1R ligand selectivity and signaling.
- Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC 3052, Australia.
Organizational Affiliation: