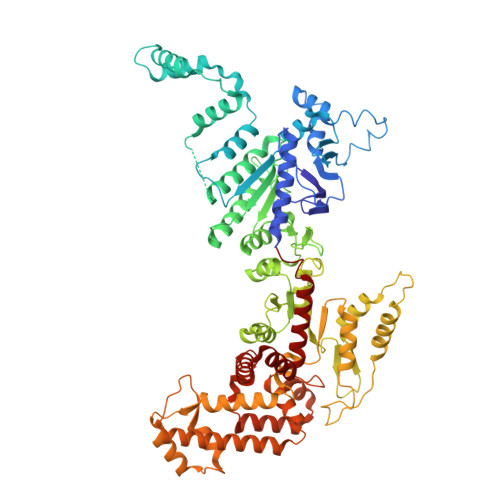
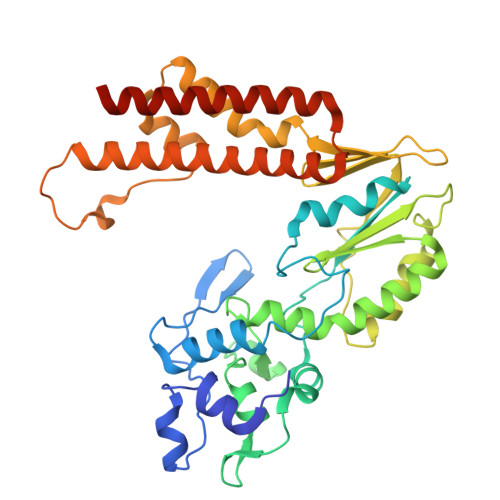
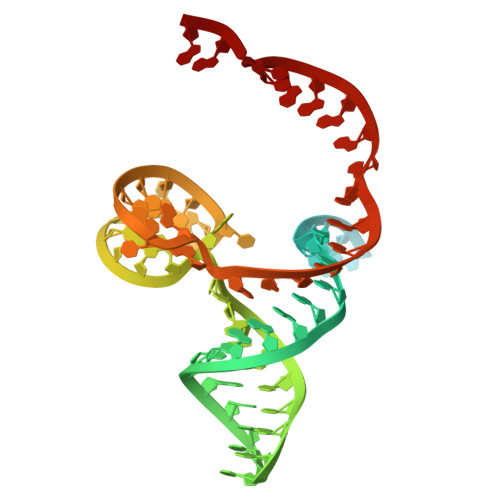
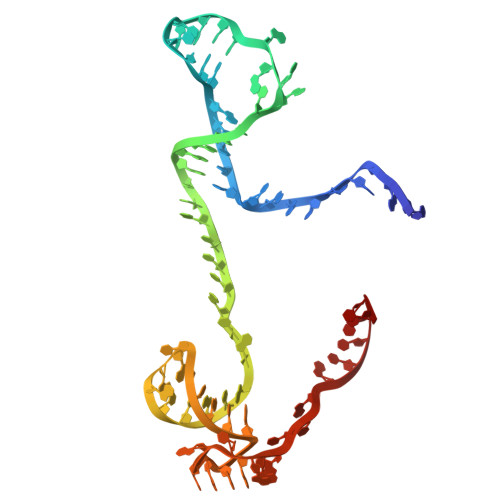
Phage SSB detection by retron Eco8 msDNA unleashes nuclease-mediated immunity.
Yu, C., Wang, C., Forman, T., Xie, J., Major, S., Fang, M.X., Voyer, J.E., Pogliano, J., Fu, T.M.(2025) Mol Cell 85: 4243
- PubMed: 41172989
- DOI: https://doi.org/10.1016/j.molcel.2025.10.007
- Primary Citation of Related Structures:
9P4J, 9P4K - PubMed Abstract:
Retrons are antiphage defense systems that synthesize multicopy single-stranded DNA (msDNA) and are being adapted for genome engineering. The Escherichia coli retron Eco8 system comprises a reverse transcriptase (RT), an msDNA, and an overcoming lysogenization defect (OLD)-family nuclease (Eco8-OLD), reminiscent of Gabija. Here, we present the cryo-electron microscopy structure of the Eco8 supramolecular complex, a symmetric 4:4:4 assembly of RT, Eco8-OLD, and msDNA (a hybrid of msrRNA and msdDNA). The msDNA anchors RT and Eco8-OLD into a compact architecture that traps Eco8-OLD in an autoinhibited conformation. Upon phage infection, phage single-stranded DNA-binding proteins (SSBs) bind msdDNA, inducing conformational rearrangements that relieve Eco8-OLD autoinhibition and activate its non-specific DNA nuclease activity. This structural transition enables Eco8 to mount a robust antiphage response by restricting phage genome amplification. Our findings reveal a previously unrecognized mechanism of retron activation and highlight the central role of msDNA as a molecular switch in controlling retron-mediated antiphage defense.
- Department of Pathology, UMass Chan Medical School, Worcester, MA 01655, USA; RNA Therapeutics Institute, UMass Chan Medical School, Worcester, MA 01655, USA; Department of Biochemistry and Molecular Biotechnology, UMass Chan Medical School, Worcester, MA 01655, USA.
Organizational Affiliation: