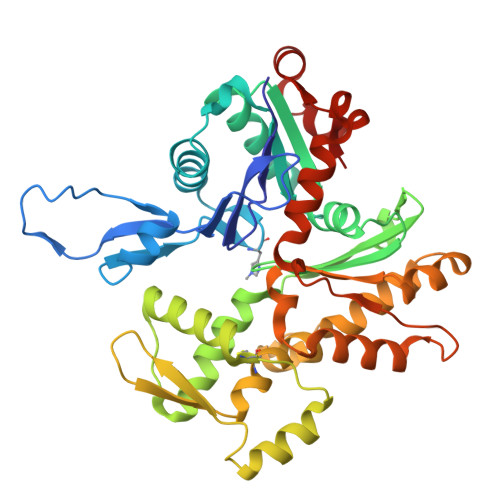
Actin isoform-specific interactions revealed by Vibrio VopV actin-binding repeats.
Kudryashova, E., Kreutzberger, M.A.B., Niedzialkowska, E., Dong, S., Kudryashov, D.S., Egelman, E.H.(2025) Proc Natl Acad Sci U S A 122: e2523856122-e2523856122
- PubMed: 41289390
- DOI: https://doi.org/10.1073/pnas.2523856122
- Primary Citation of Related Structures:
9P1I, 9P3D - PubMed Abstract:
Despite an evolutionary separation of over 300 Mya, there are no amino acid substitutions in certain actin isoforms from reptiles to mammals. What divergence that does exist between different actin isoforms is primarily tissue-specific, rather than species-specific. Sorting of actin isoforms into distinct cellular compartments is believed to be controlled by actin-binding proteins (ABPs), but little is known about how ABPs can differentiate between actin isoforms. We show that the actin-binding repeat (ABR) of the Vibrio parahaemolyticus effector VopV binds to cytoplasmic actin in a unique mode with a low nanomolar affinity, over a thousand times stronger than to muscle actin. Actin mutagenesis and cryo-EM reconstructions reveal that isoform-specific residues of previously unassigned function deep in the cleft between the two actin protofilament strands determine this selectivity. These results suggest a mechanism of highly selective, isoform-specific interactions between actin and its partners, and have broad implications for understanding the evolution of actin. Furthermore, our findings have implications in the pathogenesis of V. parahaemolyticus, whose invasion of intestinal epithelial cells relies on the interaction of VopV with cytoplasmic F-actin.
- Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210.
Organizational Affiliation: