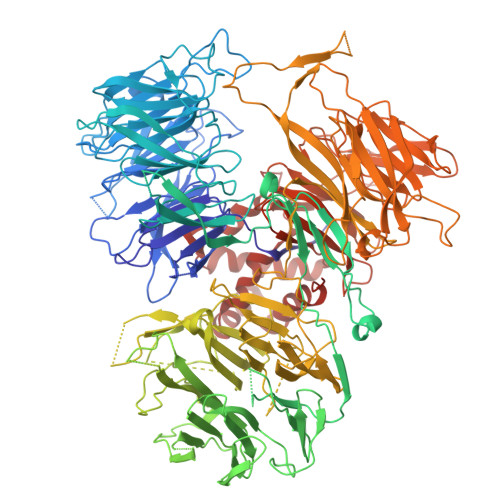
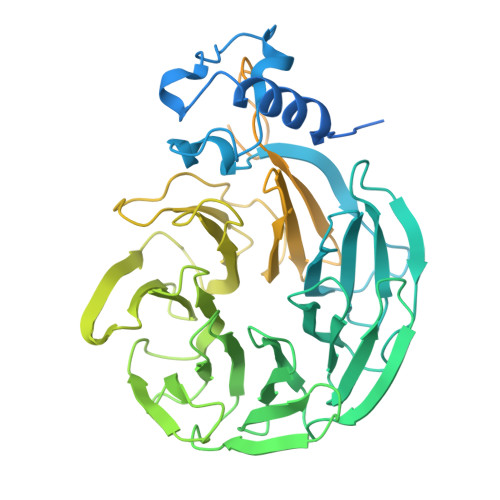
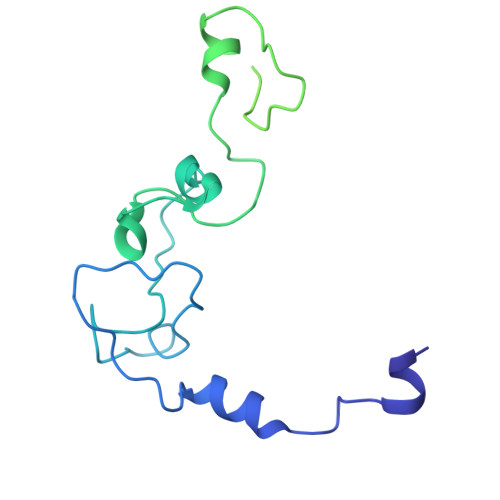
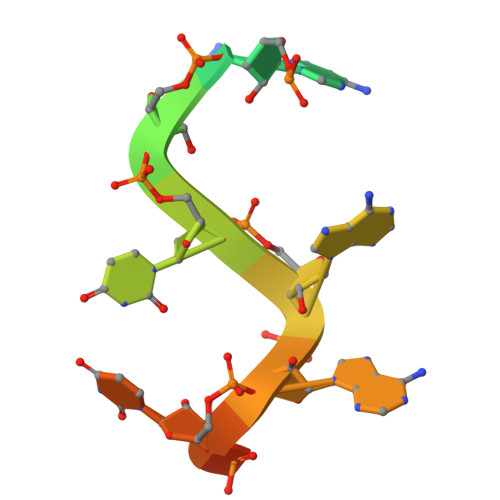
Molecular basis for the recognition of low-frequency polyadenylation signals by mPSF.
Huang, L., Chu, H.F., Tong, L.(2025) Nucleic Acids Res 53
- PubMed: 40930529
- DOI: https://doi.org/10.1093/nar/gkaf890
- Primary Citation of Related Structures:
9OXE, 9OXS - PubMed Abstract:
The 3'-end cleavage and polyadenylation of pre-mRNAs is dependent on a key hexanucleotide motif known as the polyadenylation signal (PAS). The PAS hexamer is recognized by the mammalian polyadenylation specificity factor (mPSF). AAUAAA is the most frequent PAS hexamer and together with AUUAAA, the second most frequent hexamer, account for ∼75% of the poly(A) signals. The remaining hexamers are at low frequency (<3%), and the molecular basis for their recognition is still not known. Here, we have determined the binding affinities for most of the PAS hexamers, showing that the Kd values are generally inversely correlated with their frequency. We also observed good cleavage activity for two low-frequency hexamers, AAGAAA and AACAAA. We have determined the cryo-electron microscopy structures of human mPSF in complex with AAUAAU and AGUAAA, at 3.1 and 2.5 Å resolution, respectively. The overall binding modes of the two low-frequency hexamers are similar to that of AAUAAA, although the U3-A6 Hoogsteen base pair is disrupted in the AAUAAU hexamer. For AGUAAA, the G2 base undergoes a large conformational change, which allows it to maintain the hydrogen-bonding interaction with CPSF30 as observed with A2 and establish a new hydrogen bond to CPSF30.
- Department of Biological Sciences, Columbia University, New York, NY 10027, United States.
Organizational Affiliation: