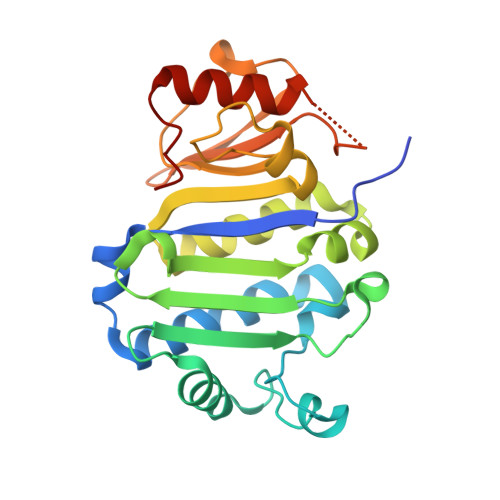
Crystal Structure of the Immunity Protein (52 domain-containing protein) from Pseudomonas aeruginosa PAO1.
Minasov, G., Wawrzak, Z., Skarina, T., Stogios, P.J., Savchenko, A., Satchell, K.F., Center for Structural Biology of Infectious Diseases (CSBID)To be published.