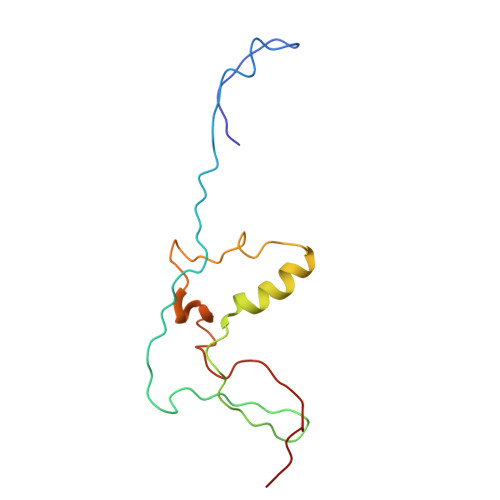
Solution Structure of the Broad-Spectrum Bacteriocin Garvicin Q.
Mallett, T., Lamer, T., Aleksandrzak-Piekarczyk, T., McKay, R.T., Catenza, K., Sit, C., Rainey, J.K., Towle-Straub, K.M., Vederas, J.C., van Belkum, M.J.(2025) Int J Mol Sci 26
- PubMed: 40869166
- DOI: https://doi.org/10.3390/ijms26167846
- Primary Citation of Related Structures:
9OIL, 9OIU - PubMed Abstract:
Class IId bacteriocins are linear, unmodified antimicrobial peptides produced by Gram-positive bacteria, and often display potent, narrow-spectrum inhibition spectra. Garvicin Q (GarQ) is a class IId bacteriocin produced by the lactic acid bacterium Lactococcus garvieae . It stands out for its unusual broad-spectrum antimicrobial activity against various bacterial species, including Listeria monocytogenes , Pediococcus pentosaceus , Carnobacterium maltaromaticum , Enterococcus faecalis , and Lactococcus spp. Its protein target is the mannose phosphotransferase system (Man-PTS) of susceptible bacterial strains, though little is known about the precise molecular mechanism behind GarQ's unusual broad spectrum of activity. In this work, 13 C- and 15 N-labelled GarQ was recombinantly produced using our previously described "sandwiched" protein expression system in Escherichia coli . We also developed a protocol to purify a uniformly labelled sample of the small ubiquitin-like modifier His 6 -SUMO, which is produced as a byproduct of the expression procedure. We demonstrated its use as a "free" protein standard for 3D NMR experiment calibrations. The GarQ solution structure was solved using triple-resonance nuclear magnetic resonance (NMR) spectroscopy and was compared with the structures of other Man-PTS-targeting bacteriocins. GarQ adopts a helix-hinge-helix fold, which is contrary to its structural predictions according to AlphaFold 3.
- Department of Chemistry, University of Alberta, Edmonton, AB T6G 2G2, Canada.
Organizational Affiliation: