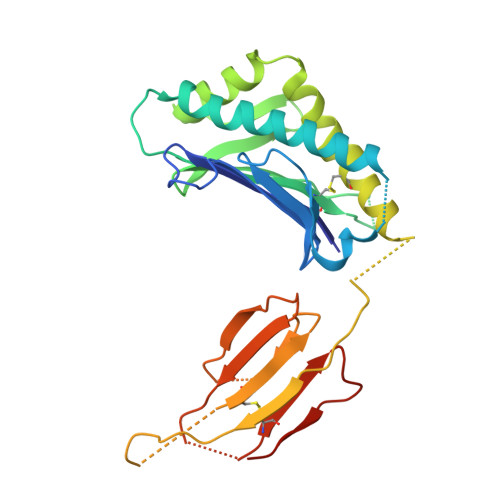
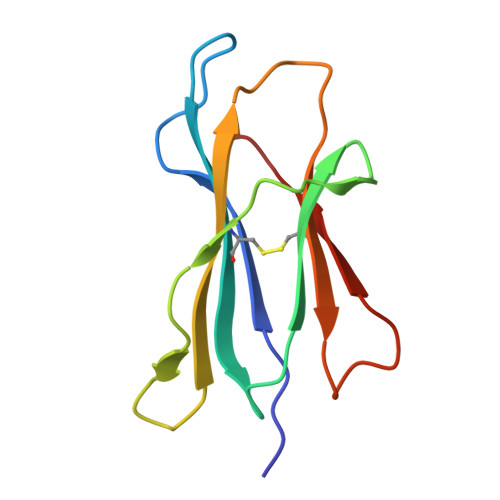
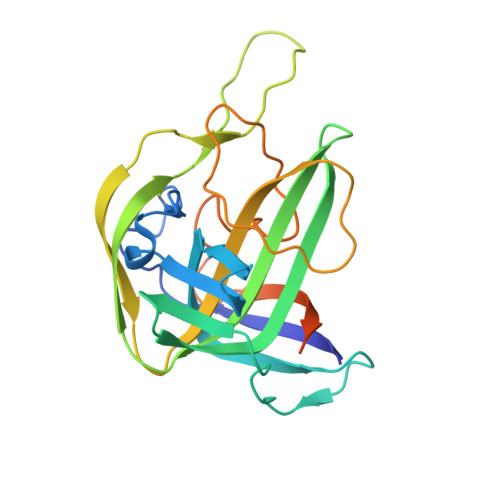
Structural hijacking of FcRn by human astrovirus spikes reveals conserved epitopes for broad-spectrum antivirals.
Agrawal, S., Jain, M., Marinelli, D., Cho, S.Y., Briney, B., Wilson, I.A.(2025) Cell Rep 44: 116679-116679
- PubMed: 41389202
- DOI: https://doi.org/10.1016/j.celrep.2025.116679
- Primary Citation of Related Structures:
9OC6, 9OC7 - PubMed Abstract:
Human astroviruses (HAstVs) are a leading cause of pediatric gastroenteritis and emerging systemic infections; however, no targeted therapies exist. A critical barrier to intervention has been the lack of molecular insights into viral entry, particularly the interaction between the HAstV capsid spike and its receptor, the neonatal Fc receptor (FcRn). Here, we report crystal structures of the HAstV spike from classical serotypes 2 and 6 in complex with human FcRn at 3 Å resolution, defining a conserved receptor-binding interface at atomic resolution. These structures reveal serotype-specific variations that dictate receptor affinity and demonstrate that reported neutralizing antibodies can inhibit infection by sterically blocking the receptor-binding site. Mapping conserved epitopes across classical HAstV serotypes provides a blueprint for designing broad-spectrum antivirals that disrupt viral entry. Notably, our structural data rationalize the potential repurposing of clinical FcRn inhibitors, such as nipocalimab, to block HAstV infection, bridging critical gaps in astrovirus biology and antiviral development.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: