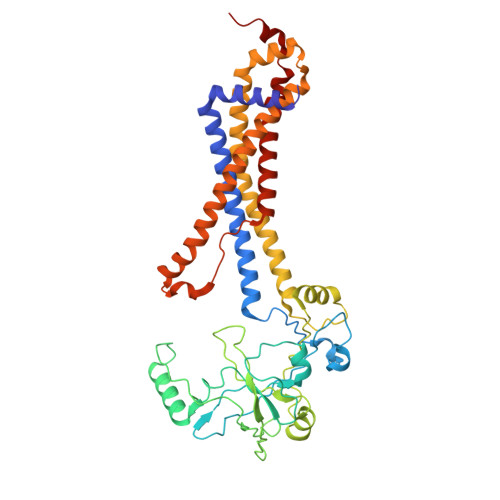
Structural analysis of a Gram-positive type VII ABC transporter induced by cell wall-targeting antibiotics.
Yu, P., Krah, B.S., Orlando, M.A., Subramanian, S., Orlando, B.J.(2026) Structure 34: 145-156.e4
- PubMed: 41161309
- DOI: https://doi.org/10.1016/j.str.2025.10.004
- Primary Citation of Related Structures:
9O9X, 9O9Y - PubMed Abstract:
Bacteria utilize a variety of mechanisms to remodel the cell wall in response to environmental and antimicrobial stress. In the model organism Bacillus subtilis, the ytr operon encoding putative ATP-binding cassette (ABC) transporter(s) is highly upregulated in response to cell wall-targeting antibiotics. Here we show that the ytr operon encodes two distinct ABC transporters: YtrBCD and YtrEF. Using cryo-electron microscopy(cryo-EM), we determined the structures of YtrEF in nucleotide-free and ADP-vanadate bound states. The structures demonstrate that YtrEF adopts a type VII ABC transporter fold. Nucleotide binding induced conformational changes that propagate from the cytosolic region through the transmembrane helices to ultimately reorient the extracellular domains. Extended bacterial growth assays and suppressor mutation identification indicated that YtrEF contributes to alteration of colony morphology. These findings establish YtrEF as a type VII ABC transporter that is induced by cell wall-targeting antibiotics and a new avenue to phenotypically assess the ytr operon.
- Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
Organizational Affiliation: