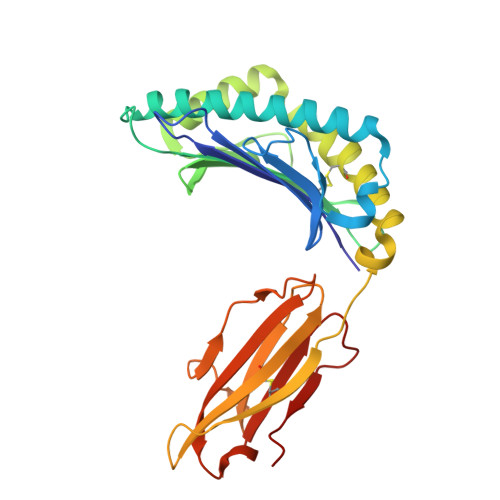
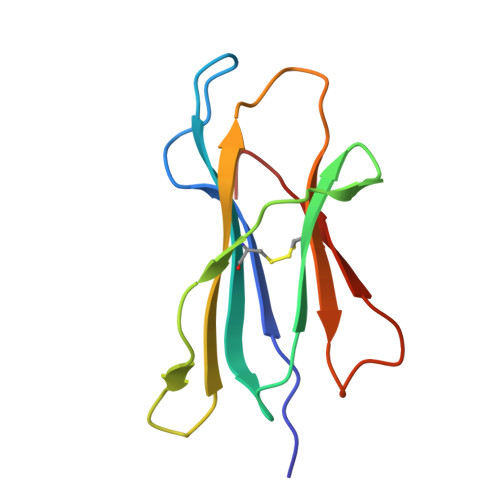
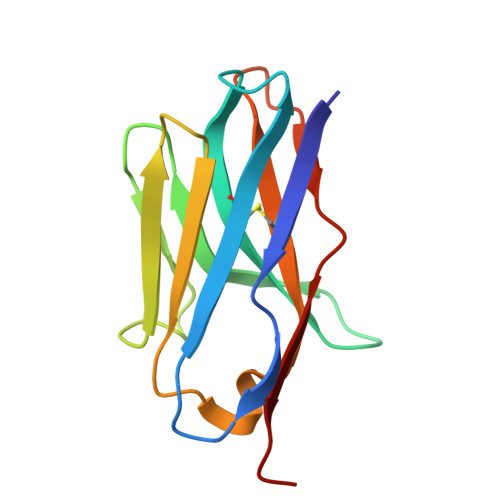
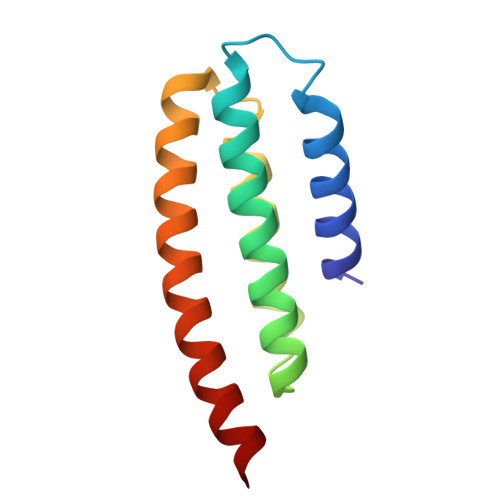
Design of high-specificity binders for peptide-MHC-I complexes.
Liu, B., Greenwood, N.F., Bonzanini, J.E., Motmaen, A., Meyerberg, J., Dao, T., Xiang, X., Ault, R., Sharp, J., Wang, C., Visani, G.M., Vafeados, D.K., Roullier, N., Nourmohammad, A., Scheinberg, D.A., Garcia, K.C., Baker, D.(2025) Science 389: 386-391
- PubMed: 40705892
- DOI: https://doi.org/10.1126/science.adv0185
- Primary Citation of Related Structures:
9O5S - PubMed Abstract:
Class I major histocompatibility complex (MHC-I) molecules present peptides derived from intracellular antigens on the cell surface for immune surveillance. Proteins that recognize peptide-MHC-I (pMHCI) complexes with specificity for diseased cells could have considerable therapeutic utility. Specificity requires recognition of outward-facing amino acid residues within the disease-associated peptide as well as avoidance of extensive contacts with ubiquitously expressed MHC. We used RFdiffusion to design pMHCI-binding proteins that make extensive contacts with the peptide and identified specific binders for 11 target pMHCs starting from either experimental or predicted pMHCI structures. Upon incorporation into chimeric antigen receptors, designs for eight targets conferred peptide-specific T cell activation. Our approach should have broad utility for both protein- and cell-based pMHCI targeting.
- Department of Biochemistry, University of Washington, Seattle, WA, USA.
Organizational Affiliation: