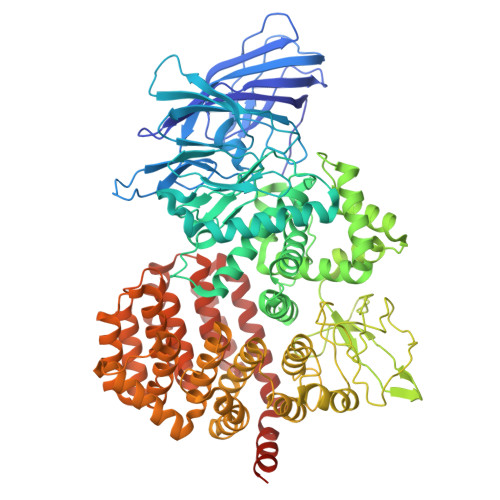
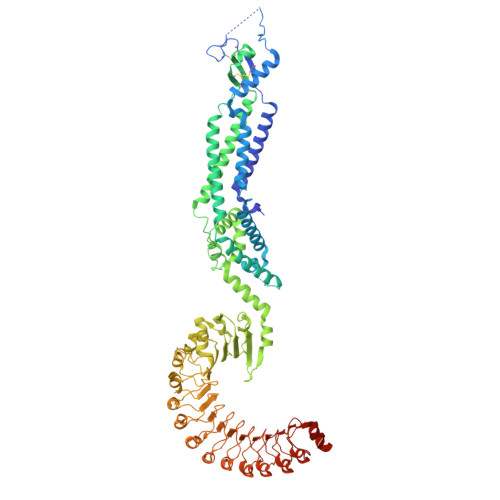
Puromycin-sensitive aminopeptidase acts as an inhibitory auxiliary subunit of volume-regulated anion channels and regulates cGAMP transport.
Zheng, W., Hagino, T., Wang, H., Cheng, H.Y., Koylass, N., Chen, K.H., Wang, H., Mani, S., Mondal, A.K., Twomey, E.C., Qiu, Z.(2025) Mol Cell 85: 4621-4632.e7
- PubMed: 41371222
- DOI: https://doi.org/10.1016/j.molcel.2025.11.014
- Primary Citation of Related Structures:
9O5K - PubMed Abstract:
Volume-regulated anion channels (VRACs) are large-pore channels expressed in most vertebrate cells and are critical for cell volume regulation and autocrine/paracrine signaling. Here, we identify the ubiquitously expressed puromycin-sensitive aminopeptidase (PSA) as a binding partner of the obligatory VRAC subunit SWELL1 (also known as LRRC8A) and determine the cryo-electron microscopy structure of the SWELL1-PSA complex. Three PSA molecules bind a single SWELL1 hexamer, coupling adjacent leucine-rich repeat (LRR) domains into local dimers. Functionally, PSA overexpression suppresses VRAC activation, whereas PSA deletion dramatically elevates basal channel activity. Notably, PSA's modulation of VRACs requires physical binding but not aminopeptidase activity, indicating a structural mechanism. Our findings identify PSA as an auxiliary subunit of VRACs, highlight the role of intracellular LRR domains in allosteric channel gating, and suggest a strategy to tune VRAC function in diverse physiological contexts, including 2'3'-cyclic GMP-AMP (cGAMP) transport and downstream stimulator of interferon genes (STING) signaling.
- Department of Physiology, Pharmacology and Therapeutics, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Organizational Affiliation: