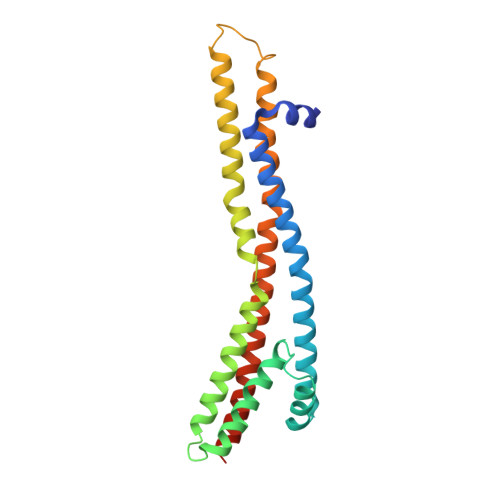
Deciphering the molecular mechanism of the bacterial division motor TolQRA.
Shen, C., Xie, T., Luo, Y., Zhao, F., Wang, X., Zhang, Z., Pang, J., Zhang, J., Dong, X., Chang, S., Ding, B.S., Ying, B., Chi, W., Su, Z., Zhou, R., Tang, X., Dong, H.(2025) Cell Discov 11: 87-87
- PubMed: 41184225
- DOI: https://doi.org/10.1038/s41421-025-00841-w
- Primary Citation of Related Structures:
9O40, 9QUQ, 9QVD - PubMed Abstract:
The Tol-Pal system is essential for maintaining outer membrane (OM) stability during cell division in Gram-negative bacteria. The inner membrane complex TolQRA harnesses proton motive force (PMF) to establish transient interactions within the periplasm, thereby coordinating cell envelope remodeling and facilitating OM invagination at division sites. However, the precise mechanism remains unclear. Here, we present cryo-electron microscopy structures of Escherichia coli TolQRA in multiple conformational states at 2.92-3.52 Å resolution, revealing rotary dynamics within the complex. Computational simulations reveal a proton-conductive channel comprising the putative proton-accepting residue Asp23 and the conserved polar residues Thr145 and Thr178, with monitored inter-residue distances providing support for a proton-driven rotary mechanism. Site-directed mutagenesis combined with functional assays validates the AlphaFold-predicted structure of the periplasmic domains of TolR and TolA, and further pinpoints critical residues required for complex function. Together, these findings advance our understanding of TolQRA-mediated proton transduction and offer new avenues for antibiotic drug development.
- Department of Laboratory Medicine, State Key Laboratory of Biotherapy, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, China.
Organizational Affiliation: