Impaired nick recognition and ligation efficiency by LIG1 K845N variant linked to Huntington's disease.
Ratcliffe, J., Lerner, C.E., Balu, K., Chatterjee, S., Lee, K.M., Caglayan, M.(2025) NAR Mol Med 2: ugaf038-ugaf038
- PubMed: 41346861
- DOI: https://doi.org/10.1093/narmme/ugaf038
- Primary Citation of Related Structures:
9NYS - PubMed Abstract:
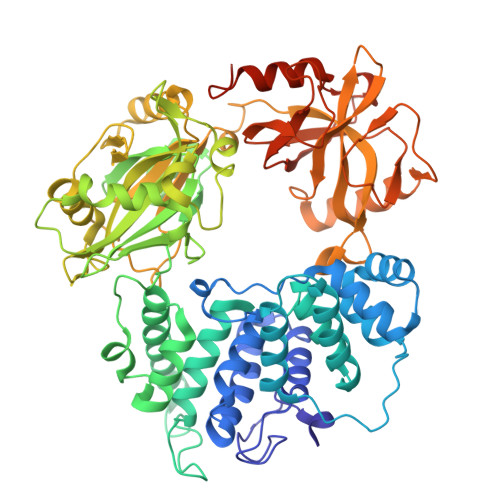
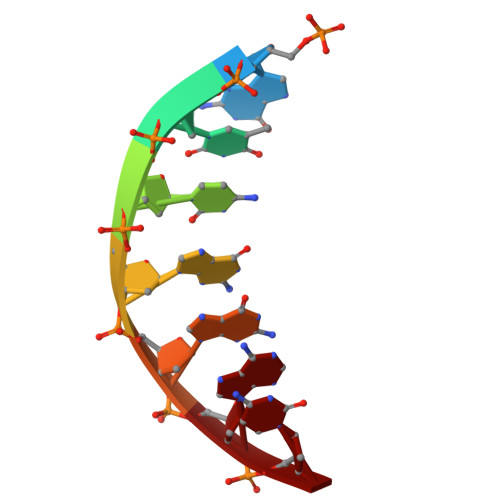
DNA ligase 1 (LIG1) joins broken strand breaks and seals Okazaki fragments during DNA repair and replication. Huntington's disease (HD)-associated mutation in the LIG1 gene, K845N, is associated with delayed symptom onset and predicted to suppress CAG repeat expansion. Yet, how this mutation impacts faithful nick sealing and efficient DNA binding by LIG1 remains unknown. Here, using biochemical analyses, X-ray crystallography, and total internal reflection fluorescence (TIRF) microscopy, we characterized the LIG1 HD-associated K845N variant at biochemical, structural, and single-molecule levels. Our results showed significantly reduced ligation efficiency for nick substrates containing noncanonical mismatches and diminished mutagenic end-joining of damaged DNA, while LIG1 K845N variant exhibits a lack of discrimination against nicks containing 3'-ribonucleotides when compared with the wild-type enzyme. Furthermore, our structures provided an atomic insight into differences in the distances between functional groups of K/N845 and DNA ends, demonstrating similar conformation at the ligase active site. Finally, our single-molecule measurements revealed that the K845N variant binds less frequently to nick, suggesting diminished affinity. Overall, our findings contribute to understanding the mechanism by which LIG1 searches for nick sites on DNA and ensures fidelity to maintain genome stability at the final ligation step in normal versus HD-associated states.
- Department of Biochemistry and Molecular Biology, University of Florida, Gainesville, FL 32610, United States.
Organizational Affiliation: