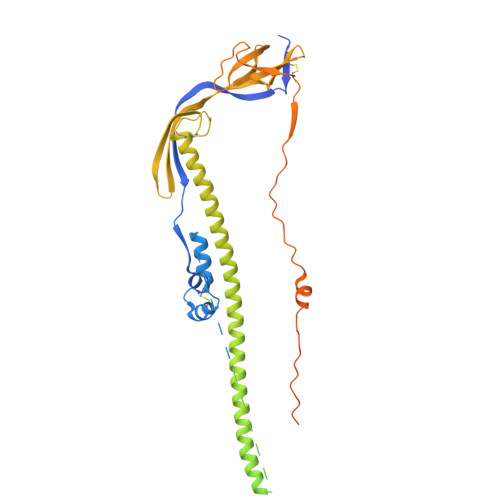
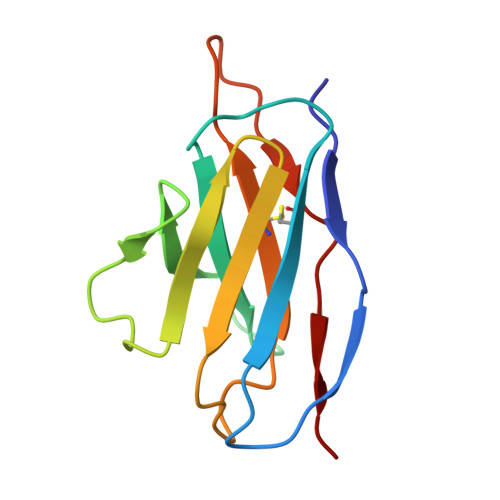
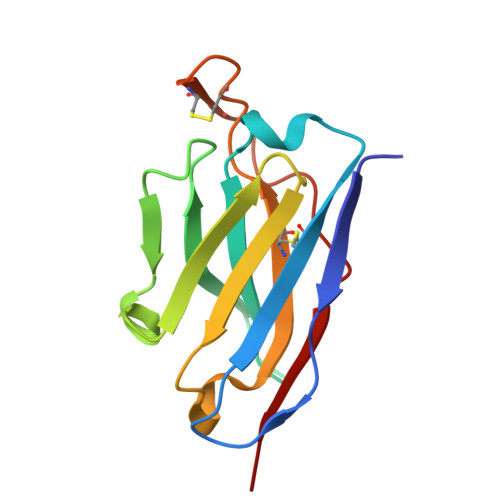
Human antibody targeting of coronavirus spike S2 subunit is associated with protection mediated by Fc effector functions.
Muthuraman, K., Jackman, M., Liang, Y., Garrett, M.E., Cui, H., Nguyen, L.V.H., Ivanochko, D., Ye, C., Pino, P.A., Hicks, A., Maingot, B., Yusko, E., Benzeno, S., Martinez-Sobrido, L., Torrelles, J.B., Gilbert, A.E., Rubin, B.E.R., Keitany, G., Jetha, A., Julien, J.-P.(2025) J Virol 99: e0152325-e0152325
- PubMed: 41222229
- DOI: https://doi.org/10.1128/jvi.01523-25
- Primary Citation of Related Structures:
9NQZ - PubMed Abstract:
Over the past two decades, betacoronaviruses (β-CoVs) have caused two epidemics and a pandemic and remain a high risk for future outbreaks through zoonotic transmissions, highlighting the need for broad biomedical countermeasures. Here, we describe a convalescent human monoclonal antibody (mAb 1871) that targets the S2 subunit of the coronavirus spike protein, with broad β-CoVs binding and sarbecovirus neutralization. Cryo-electron microscopy analysis revealed that mAb 1871 binds the upstream helix of the S2 subunit, interacting with partially conserved residues, providing a molecular basis for its cross-reactivity. Though less potent than receptor-binding domain-directed antibodies-approximately 500-fold lower neutralization potency than the emergency use authorized receptor-binding domain (RBD)-directed Pemgarda mAb against wild-type SARS-CoV-2-mAb 1871 provides protective efficacy in a mouse model. Notably, Fc effector functions are critical for its in vivo protection. This study further highlights the Fc dependence of S2-directed antibodies for in vivo protection and identifies a conserved epitope in the S2 subunit as a potential target of broad-β-CoVs countermeasures.IMPORTANCEBats and pangolins are natural reservoirs of betacoronaviruses (β-CoVs) and continue to pose a significant risk for future outbreaks through zoonotic transmissions. This highlights the need for effective countermeasures to prevent future pandemics. While neutralizing antibodies targeting the receptor-binding domain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) received emergency use authorization, many have lost efficacy as the virus evolved, and authorizations have been revoked. In contrast to the S1 subunit, the spike protein S2 subunit is more conserved across β-CoVs, making it an attractive target for the development of broadly neutralizing antibodies. Here, we describe a human mAb that targets a conserved epitope in the S2 subunit, demonstrating broad β-CoV binding, sarbecovirus neutralization, and in vivo protection mediated by Fc effector functions in a mouse model. These findings have important implications for pan-β-CoVs therapeutics and vaccine development.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Ontario, Canada.
Organizational Affiliation: