Structural basis of receptor recognition and nanobody neutralization of marburgvirus
Ye, G., Bu, F., Liu, B., Li, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanosota-MB1 | A [auth G], H, I | 158 | Vicugna pacos | Mutation(s): 0 | 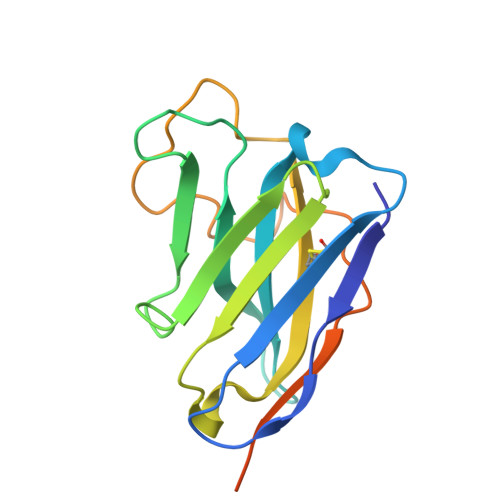 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein GP1 | B [auth A], D [auth C], F [auth E] | 258 | Ravn virus - Ravn, Kenya, 1987 | Mutation(s): 0 Gene Names: GP |  |
UniProt | |||||
Find proteins for Q1PDC7 (Lake Victoria marburgvirus (strain Ravn-87)) Explore Q1PDC7 Go to UniProtKB: Q1PDC7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q1PDC7 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein GP2 | C [auth B], E [auth D], G [auth F] | 247 | Ravn virus - Ravn, Kenya, 1987 | Mutation(s): 0 Gene Names: GP |  |
UniProt | |||||
Find proteins for Q1PDC7 (Lake Victoria marburgvirus (strain Ravn-87)) Explore Q1PDC7 Go to UniProtKB: Q1PDC7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q1PDC7 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J [auth R], K [auth S], L [auth T] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | M [auth X], N [auth Y], O [auth Z] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.1_5286: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI089728 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI171954 |