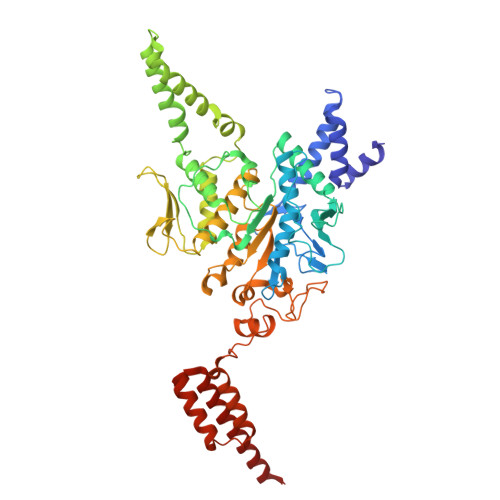
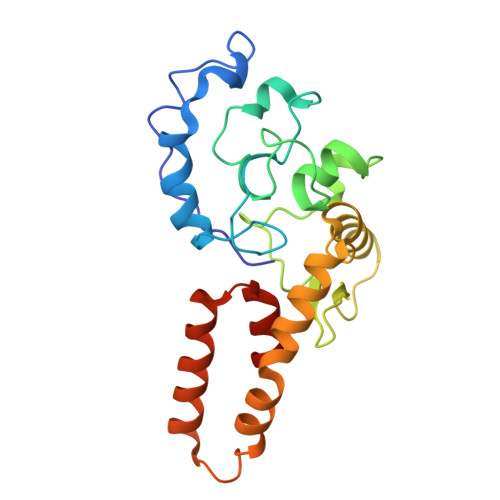
Structural basis for retron co-option of anti-phage ATPase-nuclease.
Wang, B., Hoffman, R.D., Hou, Y.M., Li, H.(2026) Nat Struct Mol Biol 33: 53-62
- PubMed: 41174277
- DOI: https://doi.org/10.1038/s41594-025-01702-6
- Primary Citation of Related Structures:
9NNB, 9NNH, 9NNK - PubMed Abstract:
Retrons have been recently identified as bacterial defense systems that employ a tripartite of reverse transcriptase, non-coding RNA (ncRNA) and its derived multi-copy single stranded DNA (msDNA) to sequester effector activity. Phage invasion activates retrons, triggering effector activity and inducing abortive infection and cell growth arrest. Ec78 differs from other retrons by leveraging the Septu defense system, a stand-alone ATPase-nuclease pair (PtuAB), by reshaping the phage sensing and molecular assembly processes of PtuAB. To elucidate how Ec78 hijacks PtuAB, we determined electron cryomicroscopy structures of Ec78 as well as the retron-displaced PtuAB. We show that the Ec78-associated ATPase, PtuA, acquired unique elements that enable its interactions with the reverse transcriptase and the msDNA, and self-assembly when displaced by the retron. By biochemical and mutational analyses, we also show that the retron-displaced PtuAB forms a tetramer, unlike its stand-alone counterpart, that restricts the host. However, in the presence of the retron, the retron-displaced PtuAB confers a well-controlled immune response, eliciting ATP hydrolysis- and msDNA-regulated targeting to host factors. Our studies reveal an evolutionary principle for retrons to co-opt conserved enzyme modules for defense in response to different cellular needs.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.
Organizational Affiliation: