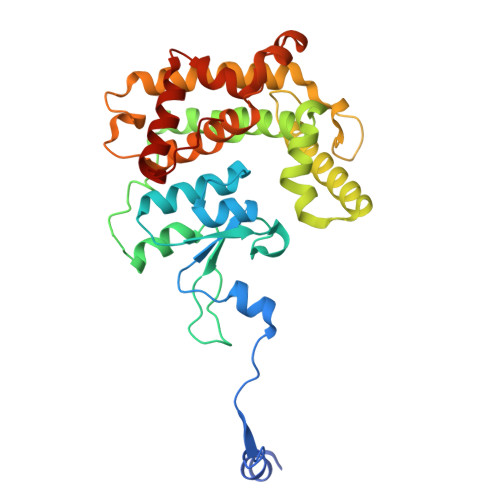
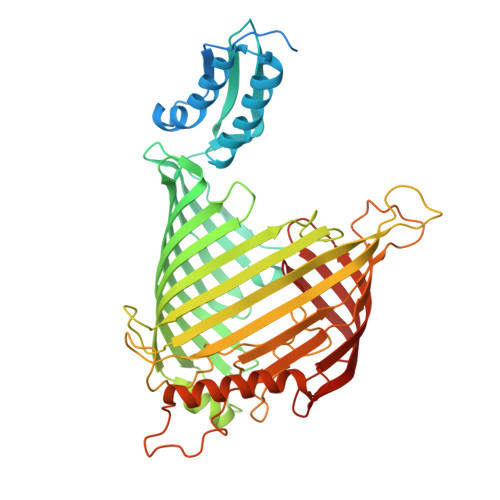
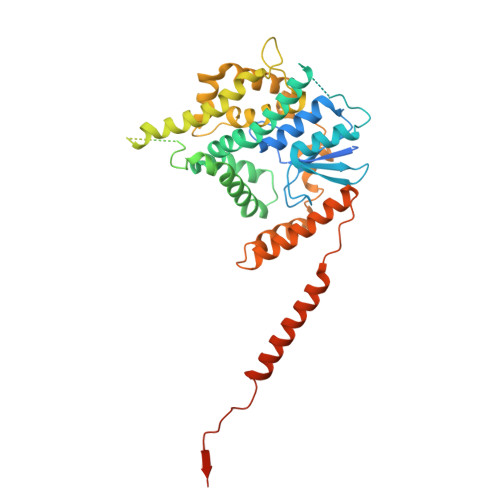
The dynamic lateral gate of the mitochondrial beta-barrel biogenesis machinery is blocked by darobactin A.
Diederichs, K.A., Botos, I., Hayashi, S., Gutishvili, G., Kotov, V., Kuo, K., Iinishi, A., Cooper, G., Schwarz, B., Celia, H., Marlovits, T.C., Lewis, K., Gumbart, J.C., Mindell, J.A., Buchanan, S.K.(2025) Nat Commun 16: 11349-11349
- PubMed: 41266328
- DOI: https://doi.org/10.1038/s41467-025-66417-0
- Primary Citation of Related Structures:
9NK6, 9NK7, 9NK8 - PubMed Abstract:
The folding and insertion of β-barrel proteins into the mitochondrial outer membrane is facilitated by the sorting and assembly machinery (SAM) complex. Here we report two 2.8 Å cryo-EM structures of the Thermothelomyces thermophilus SAM complex in the absence of substrate in which the Sam50 lateral gate adopts two different conformations: the first is a closed lateral gate as observed in previously published structures, while the second contains a Sam50 with the first four β-strands rotated outwards by approximately 45°, resulting in an open lateral gate. The observed monomeric open conformation contrasts our previous work where the open conformation was adopted by non-physiological up-down dimers. To understand how these lateral gate dynamics are influenced by substrate, we studied the interaction of the SAM complex with a β-signal peptide mimic, darobactin A. Darobactin A binds to the SAM complex with nanomolar affinity and inhibits the import and assembly of mitochondrial β-barrel proteins in vitro. Lastly, we solved a 3.0 Å cryo-EM structure of the Thermothelomyces thermophilus SAM complex bound to darobactin A, which reveals that darobactin A stabilizes the Sam50 lateral gate similar to the open conformation by binding to strand β1, therefore blocking β-barrel biogenesis.
- Laboratory of Molecular Biology, National Institute of Diabetes & Digestive & Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: