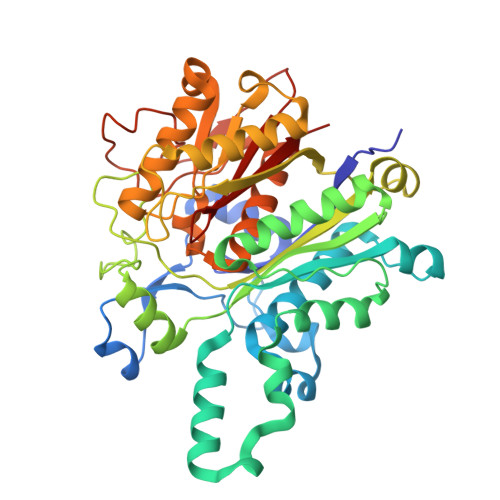
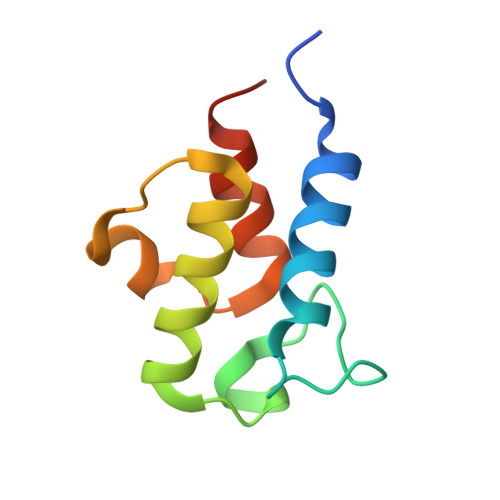
Role of Human Mitochondrial Ketosynthase in Long-Chain Fatty Acid Biosynthesis.
Suo, Y., Jiang, Z., Heberlig, G.W., Wang, E.Y., Sankaran, B., La Clair, J.J., Burkart, M.D.(2025) J Am Chem Soc 147: 33248-33255
- PubMed: 40891531
- DOI: https://doi.org/10.1021/jacs.5c10318
- Primary Citation of Related Structures:
9N50, 9N51 - PubMed Abstract:
Although ketosynthases in bacterial type II fatty acid biosynthesis have been extensively studied, the human mitochondrial ketosynthase, OXSM, remains incompletely characterized. Contrary to the assumption that the role of mitochondrial fatty acid biosynthesis is limited to the production of the lipoic acid precursor octanoate, recent studies suggest an ability to produce longer chain fatty acids. Here, we employ covalent, dual site-selective cross-linkers to trap the interactions between OXSM and its mitochondrial acyl carrier protein partner, mACP. Two high-resolution crystal structures that capture OXSM bound to mACP provide molecular details of the conformational changes that guide the chain elongation process. We identify key protein • protein and protein • substrate interactions that regulate the transacylation and condensation steps associated with this process. We also observe a conserved gating mechanism previously identified in bacterial type II ketosynthases. Complemented by site-directed mutagenesis and activity analyses, these findings provide detailed insight into the selectivity of the OXSM substrate. This study explores how the OXSM can elongate fatty acids larger than eight carbons, mirroring that of its bacterial type II progenitor.
- Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92037-0358, United States.
Organizational Affiliation: