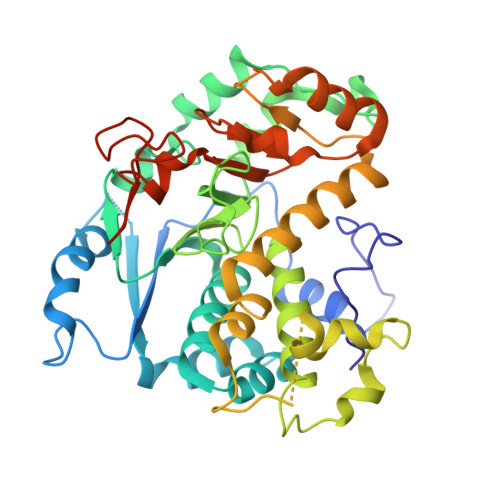
Crystal structure of the fungal mannosyltransferase Och1 reveals active site primed for N-glycan binding.
Kelly, E.T.R., Rodionov, D., Sleno, B., Romero, P.A., Berghuis, A.M.(2025) PLoS One 20: e0329259-e0329259
- PubMed: 40743235
- DOI: https://doi.org/10.1371/journal.pone.0329259
- Primary Citation of Related Structures:
9N3S - PubMed Abstract:
The outermost layer of a fungi's cell wall serves as the organism's point of first contact with its environment, or host. Heavily glycosylated glycoproteins anchor a complex meshwork of branching mannose chains, forming the outer cell wall layer in most yeast and mold species. Outer mannan chains are composed of large polymannose branching glycans attached to the universal eukaryotic N-glycan GlcNAc2Man8 core. Synthesized in the endoplasmic reticulum, the core N-glycan is transferred to the Golgi apparatus, where the first fungi-specific reaction takes place. In the cis-Golgi, Och1 (Outer chain elongation 1) plays a central role in initiating outer mannan cell wall synthesis by transferring a single α-1,6-mannose residue to the N-GlcNAc2Man8 core. Playing a vital role in fungal biology, fungal cell wall synthesis proteins have long since been thought as attractive options in the search for a fungi-specific drug target. Saccharomyces cerevisiae Δ52-Och1 was expressed in Pichia pastoris. Here, the first X-ray crystal structure of a fungal Och1 protein is reported, determined to 2.0 Å. Molecular modeling of ligand binding and sequence analysis has revealed a highly conserved substrate binding site, rationalizing Och1 target specificity for the N-GlcNAc2Man8 glycan.
- Department of Biochemistry, McGill University, Montréal, Québec, Canada.
Organizational Affiliation: