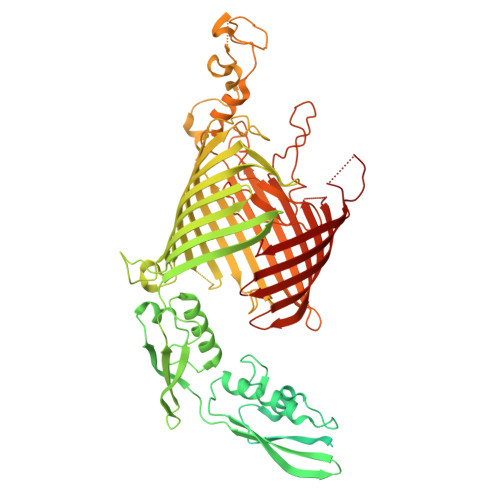
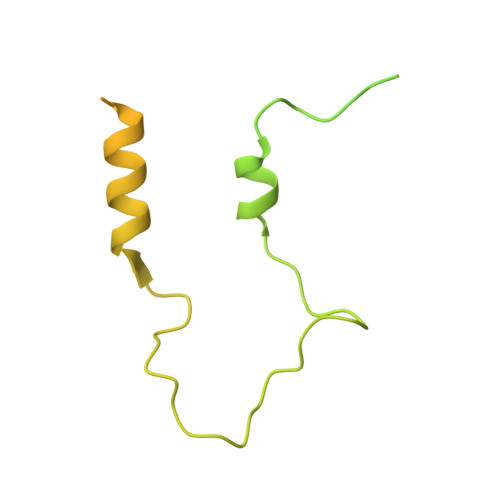
A shared mechanism for Bacteroidota protein transport and gliding motility.
Liu, X., Avramova, M., Deme, J.C., Jones, R.L., Lundgren, C.A.K., Lea, S.M., Berks, B.C.(2025) Nat Commun 16: 10217-10217
- PubMed: 41266322
- DOI: https://doi.org/10.1038/s41467-025-65003-8
- Primary Citation of Related Structures:
9N2D, 9N2E - PubMed Abstract:
Bacteria of the phylum Bacteroidota are major human commensals and pathogens in addition to being abundant members of the wider biosphere. Bacteroidota move by gliding and they export proteins using the Type 9 Secretion System (T9SS). Here we discover that gliding motility and the T9SS share an unprecedented mechanism of energisation in which outer membrane proteins are covalently attached by disulfide bonds to a moving internal track structure that propels them laterally through the membrane. We determined the structure of an exemplar Bacteroidota mobile track by obtaining the cryoEM structure of a 3 MDa circular mini-track from Porphyromonas gingivalis. Our discoveries identify a mechanistic and evolutionary link between gliding motility and T9SS-dependent protein transport.
- Department of Biochemistry, University of Oxford, Oxford, UK. xiaolong.liu@bioch.ox.ac.uk.
Organizational Affiliation: