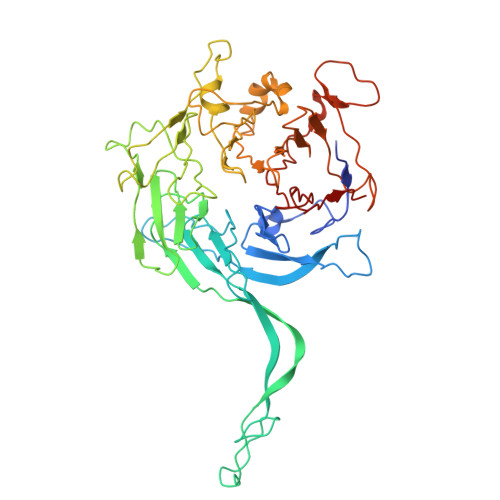
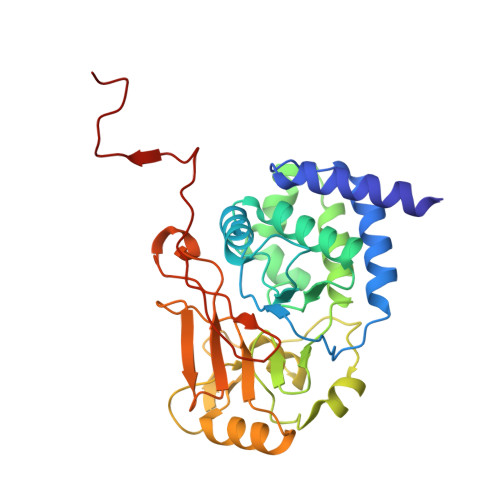
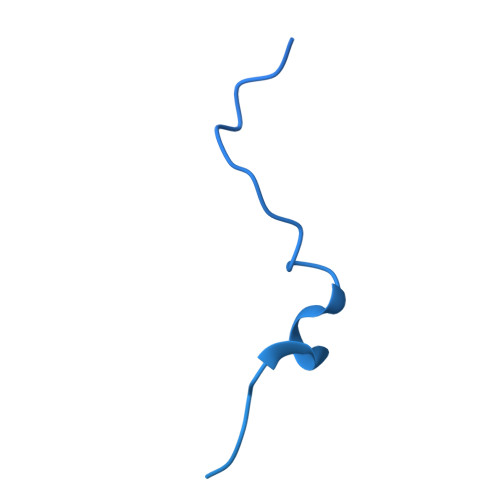
Cryo-EM structures reveal the PP2A-B55 alpha and Eya3 interaction that can be disrupted by a peptide inhibitor.
Shi, S., Li, X., Alderman, C., Wick, L., Huang, W., Foulon, N., Zhang, L., Rossi, J., Hu, W., Cui, S., Zheng, H., Taylor, D.J., Ford, H.L., Zhao, R.(2025) J Biological Chem 301: 110287-110287
- PubMed: 40414499
- DOI: https://doi.org/10.1016/j.jbc.2025.110287
- Primary Citation of Related Structures:
9N0Y, 9N0Z - PubMed Abstract:
We have previously shown that Eya3 recruits PP2A-B55α to dephosphorylate pT58 on Myc, increasing Myc stability and enhancing primary tumor growth of triple-negative breast cancer (TNBC). However, the molecular details of how Eya3 recruits PP2A-B55α remain unclear. Here we determined the cryo-EM structures of PP2A-B55α bound with Eya3, with an inhibitory peptide B55i, and in its unbound state. These studies demonstrate that Eya3 binds B55α through an extended peptide in the NTD of Eya3. The Eya3 peptide, PP2A-B55α substrates, and protein/peptide inhibitors including B55i bind to a similar area on the B55α surface but the molecular details of the binding differ. We further demonstrated that the B55i peptide inhibits the B55α and Eya3 interaction in vitro. The B55i peptide expressed on a plasmid increases Myc pT58 and decreases Myc protein levels in TNBC cells, suggesting the potential of B55i or similar peptides as therapies for TNBC.
- Department of Biochemistry and Molecular Genetics, University of Colorado Anschutz Medical Campus, Aurora, CO 80045.
Organizational Affiliation: