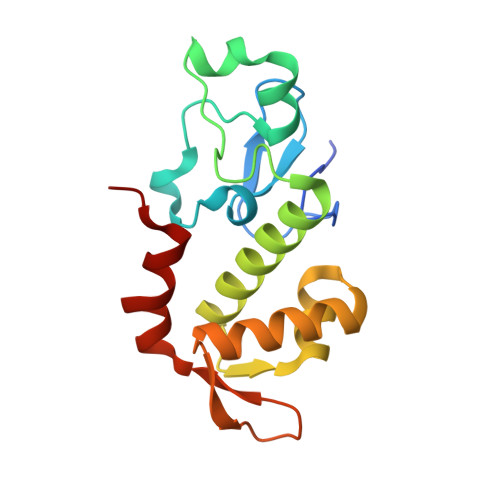
Bipartite chromatin recognition by Hop1 from two diverged Holozoa.
Rodriguez, A.A., Cirulli, A.E., Chau, K., Nguyen, J., Ye, Q., Corbett, K.D.(2025) Life Sci Alliance 8
- PubMed: 40829932
- DOI: https://doi.org/10.26508/lsa.202503428
- Primary Citation of Related Structures:
9MUG, 9MYP - PubMed Abstract:
In meiosis, ploidy reduction is driven by a complex series of DNA breakage and recombination events between homologous chromosomes, orchestrated by meiotic HORMA domain proteins (HORMADs). Meiotic HORMADs possess a central chromatin binding region (CBR) whose architecture varies across eukaryotic groups. Here, we determine high-resolution crystal structures of the meiotic HORMAD CBR from two diverged aquatic Holozoa, Schistosoma mansoni and Patiria miniata , which reveal tightly associated plant homeodomain (PHD) and winged helix-turn-helix (wHTH) domains. We show that PHD-wHTH CBRs bind duplex DNA through their wHTH domains, and identify key residues that disrupt this interaction. Combining experimental and predicted structures, we show that the CBRs' PHDs likely interact with the tail of histone H3, and may discriminate between unmethylated and trimethylated H3 lysine 4. Finally, we show that Holozoa Hop1 CBRs bind nucleosomes in vitro in a bipartite manner involving both the PHD and wHTH domain. Our data reveal how meiotic HORMADs with PHD-wHTH CBRs can bind chromatin and potentially discriminate between chromatin states to drive meiotic recombination to specific chromosomal regions.
- Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA, USA.
Organizational Affiliation: