Directed evolution of alpha-ketoisovalerate decarboxylase for improved isobutanol and 3-methyl-1-butanol production in cyanobacteria.
Xie, H., Begum, A., Gunn, L.H., Lindblad, P.(2025) Biotechnol Biofuels Bioprod 18: 84-84
- PubMed: 40745552
- DOI: https://doi.org/10.1186/s13068-025-02687-6
- Primary Citation of Related Structures:
9MSA - PubMed Abstract:
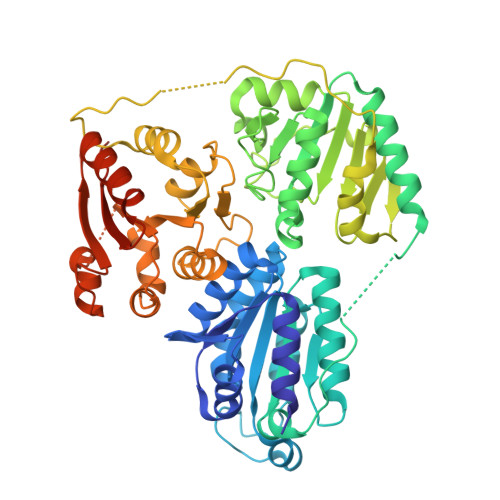
Cyanobacteria are promising platforms for metabolic engineering to convert carbon dioxide into valuable fuels and chemicals, addressing both energy demands and global climate change. Among various fuels and chemicals, isobutanol (IB) and 3-methyl-1-butanol (3M1B) have gained increasing attention due to their superior physical properties, such as high energy density, low water solubility, and low hygroscopicity. Heterologously expressing α-ketoisovalerate decarboxylase (Kivd S286T ) in the unicellular cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis) enables microbial production of IB and 3M1B through the 2-keto acid pathway, with Kivd S286T identified as a key bottleneck limiting production efficiency. To address this limitation, a high-throughput screening method based on the consumption of the substrate 2-ketoisovalerate was successfully established. This screen was coupled with random mutagenesis, via error-prone PCR, of Kivd S286T . Out of the 1600 variants, 1B12, featuring dual substitutions K419E and T186S, exhibited a 55% increase in IB production and a 50% increase in 3M1B production in Synechocystis on the fourth day of cultivation. The crystal structure of Kivd S286T was determined as a tetramer with a resolution of 2.8 Å to provide a framework for analyzing the structural basis for the enhanced butanol production conferred by the K419E and T186S substitutions. A novel Kivd variant, 1B12, was successfully generated via directed evolution, with enhanced catalytic activity for microbial IB and 3M1B biosynthesis. To our knowledge, this study represents the first successful application of directed evolution on the rate-limiting enzyme of a specific metabolic pathway to enhance biochemical production in cyanobacteria.
- Microbial Chemistry, Department of Chemistry-Ångström Laboratory, Uppsala University, Uppsala, Sweden.
Organizational Affiliation: