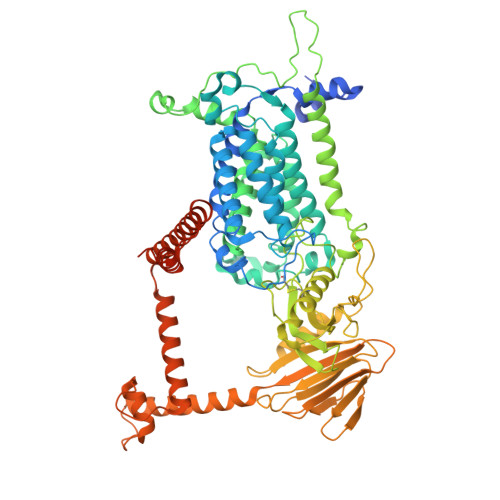
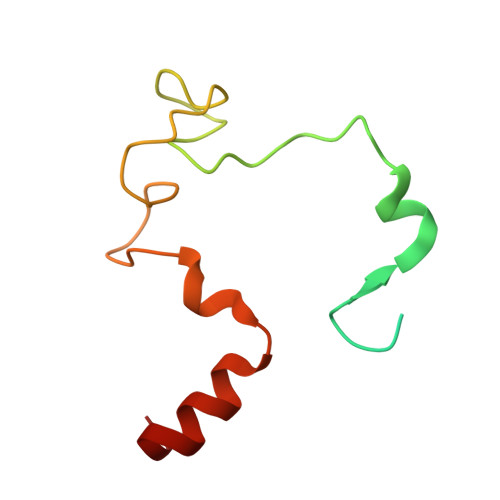
Structural insights into the vitamin K-dependent gamma-carboxylation of osteocalcin.
Cao, Q., Fan, J., Ammerman, A., Awasthi, S., Lin, Z., Mierxiati, S., Chen, H., Xu, J., Garcia, B.A., Liu, B., Li, W.(2025) Cell Res 35: 735-749
- PubMed: 40890294
- DOI: https://doi.org/10.1038/s41422-025-01161-0
- Primary Citation of Related Structures:
9MQB, 9MQC, 9MQE - PubMed Abstract:
The γ-carboxylation state of osteocalcin determines its essential functions in bone mineralization or systemic metabolism and serves as a prominent biomarker for bone health and vitamin K nutrition. This post-translational modification of glutamate residues is catalyzed by the membrane-embedded vitamin K-dependent γ-carboxylase (VKGC), which typically recognizes protein substrates through their tightly bound propeptide that triggers γ-carboxylation. However, the osteocalcin propeptide exhibits negligible affinity for VKGC. To understand the underlying molecular mechanism, we determined the cryo-electron microscopy structures of VKGC with osteocalcin carrying a native propeptide or a high-affinity variant at different carboxylation states. The structures reveal a large chamber in VKGC that maintains uncarboxylated and partially carboxylated osteocalcin in partially unfolded conformations, allowing their glutamate-rich region and C-terminal helices to engage with VKGC at multiple sites. Binding of this mature region together with the low-affinity propeptide effectively stimulates VKGC activity, similar to high-affinity propeptides that differ only in closely fitting interactions. However, the low-affinity propeptide renders osteocalcin prone to undercarboxylation at low vitamin K levels, thereby serving as a discernible biomarker. Overall, our studies reveal the unique interaction of osteocalcin with VKGC and provide a framework for designing therapeutic strategies targeting osteocalcin-related bone and metabolic disorders.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO, USA.
Organizational Affiliation: