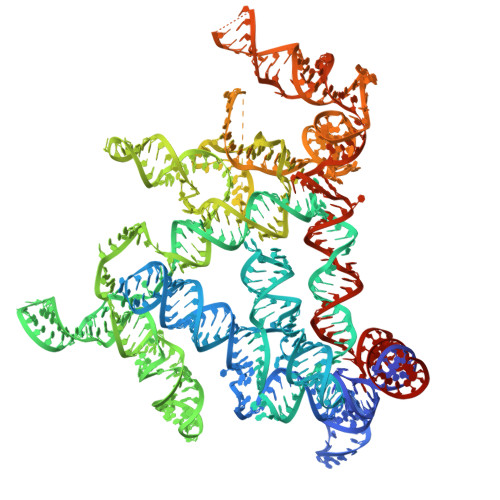
Cryo-EM structure of a natural RNA nanocage.
Ling, X., Golovenko, D., Gan, J., Ma, J., Korostelev, A.A., Fang, W.(2025) Nature 644: 1107-1115
- PubMed: 40523399
- DOI: https://doi.org/10.1038/s41586-025-09262-x
- Primary Citation of Related Structures:
9MM6, 9MME, 9MMG - PubMed Abstract:
Long (>200 nucleotides) non-coding RNAs (lncRNAs) play important roles in diverse aspects of life. Over 20 classes of lncRNAs have been identified in bacteria and bacteriophages through comparative genomics analyses, but their biological functions remain largely unexplored 1-3 . Due to the large sizes, the structural determinants of most lncRNAs also remain uncharacterized. Here we report the structures of two natural RNA nanocages formed by the lncRNA ROOL (rumen-originating, ornate, large) found in bacterial and phage genomes. ~2.9 Å cryo-electron microscopy (cryo-EM) structures reveal that ROOL RNAs form an octameric nanocage with a 28-nm diameter and 20-nm axial length, whose hollow inside features poorly ordered regions. The octamer is stabilized by numerous tertiary and quaternary interactions, including triple-strand A-minors that we propose to name "A-minor staples". The structure of an isolated ROOL monomer at ~3.2-Å resolution indicates that nanocage assembly involves a strand-swapping mechanism resulting in quaternary kissing loops. Finally, we show that ROOL RNA fused to an RNA aptamer, tRNA, or microRNA retains its structure forming a nanocage with radially displayed cargos. Our findings therefore may enable the engineering of novel RNA nanocages as delivery vehicles for research and therapeutic applications.
- RNA Therapeutics Institute, UMass Chan Medical School, Worcester, MA, USA. xiaobin.ling@umassmed.edu.
Organizational Affiliation: