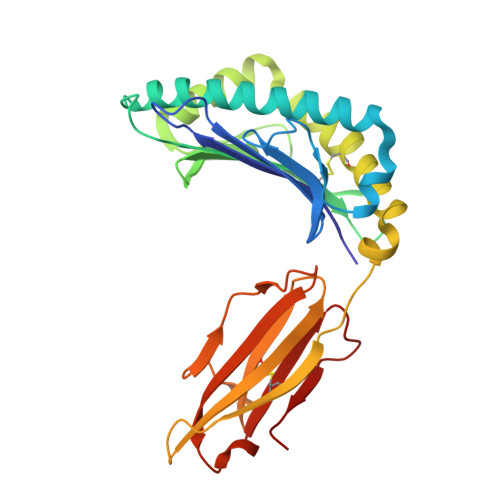
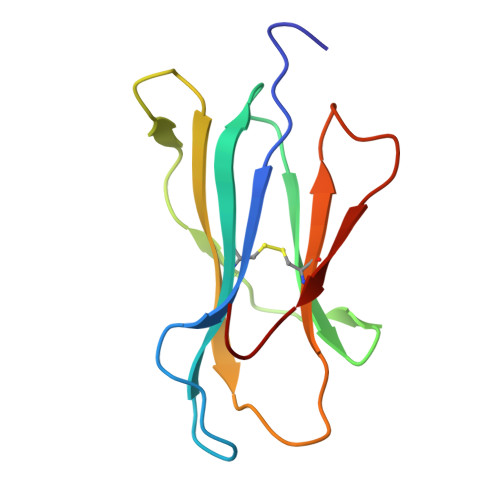
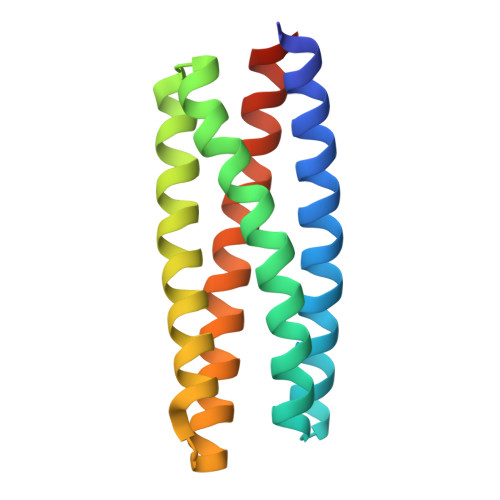
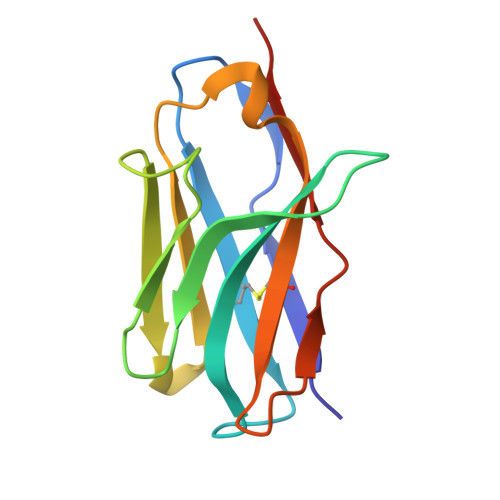
De novo design and structure of a peptide-centric TCR mimic binding module.
Householder, K.D., Xiang, X., Jude, K.M., Deng, A., Obenaus, M., Zhao, Y., Wilson, S.C., Chen, X., Wang, N., Garcia, K.C.(2025) Science 389: 375-379
- PubMed: 40705894
- DOI: https://doi.org/10.1126/science.adv3813
- Primary Citation of Related Structures:
9MIN - PubMed Abstract:
T cell receptor (TCR) mimics offer a promising platform for tumor-specific targeting of peptide-major histocompatibility complex (pMHC) in cancer immunotherapy. In this study, we designed a de novo α-helical TCR mimic (TCRm) specific for the NY-ESO-1 peptide presented by human leukocyte antigen (HLA)-A*02, achieving high on-target specificity with nanomolar affinity (dissociation constant K d = 9.5 nM). The structure of the TCRm-pMHC complex at 2.05-Å resolution revealed a rigid TCR-like docking mode with an unusual degree of focus on the up-facing NY-ESO-1 side chains, suggesting the potential for reduced off-target reactivity. Indeed, a structure-informed in silico screen of 14,363 HLA-A*02 peptides correctly predicted two off-target peptides, yet our TCRm maintained peptide selectivity and cytotoxicity as a T cell engager. These results represent a path for precision targeting of tumor antigens with peptide-focused α-helical TCR mimics.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.
Organizational Affiliation: