Cryo-EM structure of violaxanthin-chlorophyll a protein with red Chl: rVCP from Trachydiscus minutus.
Seki, S., Litvin, R., Bina, D., Miyata, T., Tanaka, H., Namba, K., Kurisu, G., Polivka, T., Fujii, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Light-harvesting complex | 229 | Trachydiscus minutus | Mutation(s): 0 | 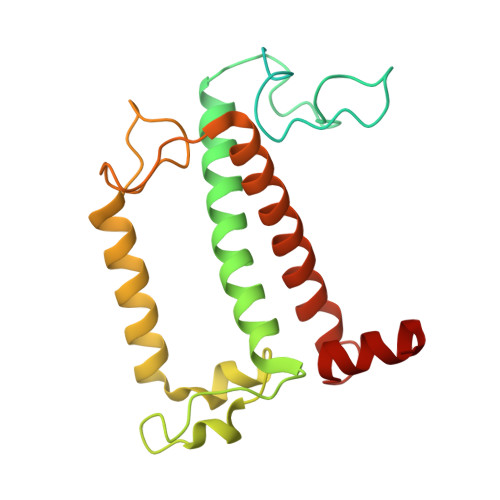 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| light-harvesting complex | 221 | Trachydiscus minutus | Mutation(s): 0 | 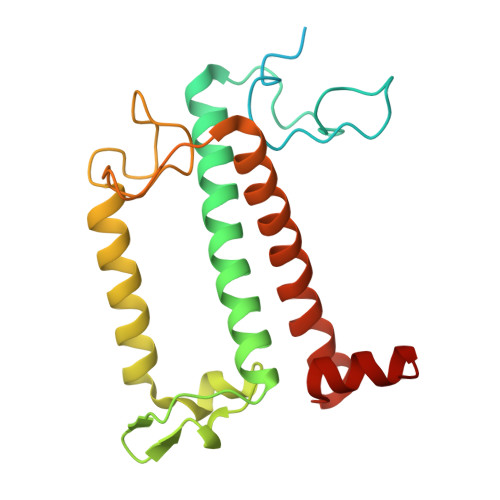 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CLA (Subject of Investigation/LOI) Query on CLA | AA [auth B] AB [auth D] BA [auth B] BB [auth D] CA [auth B] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| A1L1F Query on A1L1F | FA [auth B] KB [auth D] Q [auth A] S [auth A] VA [auth C] | [(2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E})-17-[(4~{S},6~{R})-4-acetyloxy-2,2,6-trimethyl-6-oxidanyl-cyclohexylidene]-6,11,15-trimethyl-2-[(~{E})-2-[(1~{S},4~{S},6~{R})-2,2,6-trimethyl-4-oxidanyl-7-oxabicyclo[4.1.0]heptan-1-yl]ethenyl]heptadeca-2,4,6,8,10,12,14,16-octaenyl] octanoate C50 H72 O7 JYJFYJLLQQTZBJ-AYSGSIDZSA-N |  | ||
| XAT Query on XAT | GA [auth B] HA [auth B] IA [auth B] LB [auth D] MB [auth D] | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL C40 H56 O4 SZCBXWMUOPQSOX-WVJDLNGLSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | Coot | 0.9.4.1 |
| MODEL REFINEMENT | PHENIX | 1.19.2 |
| RECONSTRUCTION | cryoSPARC | 4.2.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Agency for Medical Research and Development (AMED) | Japan | JP23ama121003 |
| Japan Society for the Promotion of Science (JSPS) | Japan | K23KJ1834 |
| Japan Science and Technology | Japan | JPMJFS2138 |
| Japan Science and Technology | Japan | JPMJCR20E1 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 23H04958 |