Cryo-EM structure of the Dinoroseobacter shibae RC-LH1 supercomplex
Liu, Z.K., Wang, P., Liu, L.N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein LRC | 76 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 | 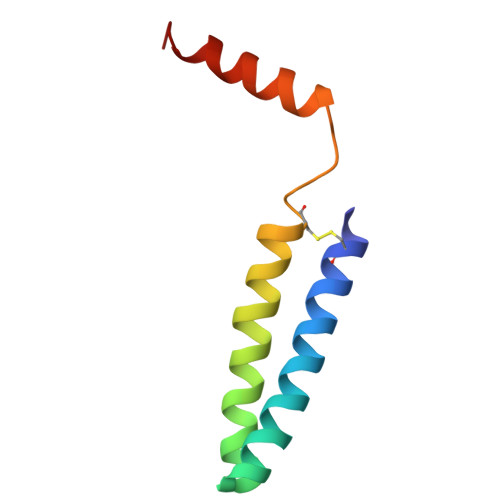 | |
UniProt | |||||
Find proteins for A8LIU2 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LIU2 Go to UniProtKB: A8LIU2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LIU2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antenna pigment protein alpha chain | 51 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 | 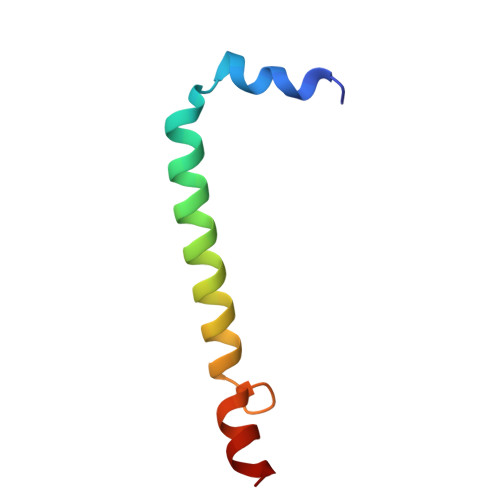 | |
UniProt | |||||
Find proteins for A8LQ15 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ15 Go to UniProtKB: A8LQ15 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ15 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antenna pigment protein beta chain | 44 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 | 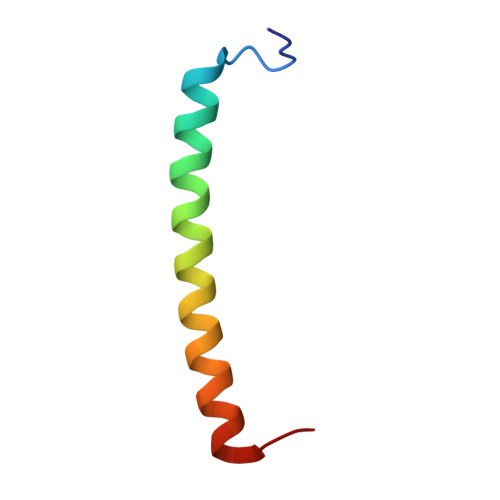 | |
UniProt | |||||
Find proteins for A8LQ14 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ14 Go to UniProtKB: A8LQ14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosynthetic reaction center cytochrome c subunit | HA [auth C] | 357 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 | 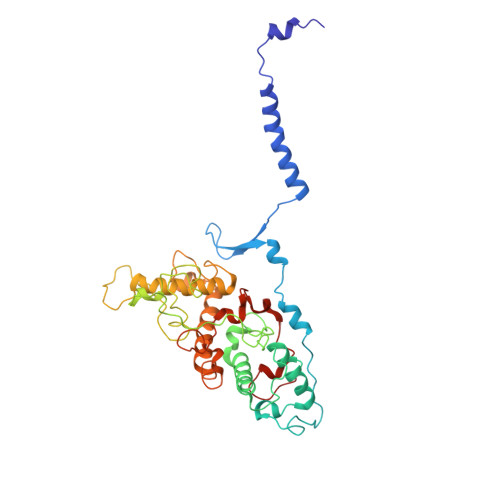 |
UniProt | |||||
Find proteins for A8LQ18 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ18 Go to UniProtKB: A8LQ18 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ18 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein H chain | IA [auth H] | 255 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A8LQ33 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ33 Go to UniProtKB: A8LQ33 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ33 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein L chain | JA [auth L] | 273 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A8LQ16 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ16 Go to UniProtKB: A8LQ16 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ16 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein M chain | KA [auth M] | 325 | Dinoroseobacter shibae DFL 12 = DSM 16493 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A8LQ17 (Dinoroseobacter shibae (strain DSM 16493 / NCIMB 14021 / DFL 12)) Explore A8LQ17 Go to UniProtKB: A8LQ17 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A8LQ17 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BCL (Subject of Investigation/LOI) Query on BCL | AB [auth 4] AC [auth l] AE [auth 0] BB [auth w] BD [auth 8] | BACTERIOCHLOROPHYLL A C55 H74 Mg N4 O6 DSJXIQQMORJERS-AGGZHOMASA-M |  | ||
| BPH (Subject of Investigation/LOI) Query on BPH | LD [auth L], SD [auth M], WD [auth M] | BACTERIOPHEOPHYTIN A C55 H76 N4 O6 KWOZSBGNAHVCKG-SZQBJALDSA-N |  | ||
| U10 (Subject of Investigation/LOI) Query on U10 | KD [auth L], NA [auth A], ND [auth L], QD [auth M] | UBIQUINONE-10 C59 H90 O4 ACTIUHUUMQJHFO-UPTCCGCDSA-N |  | ||
| MW9 (Subject of Investigation/LOI) Query on MW9 | DC [auth g] EC [auth g] FD [auth H] GD [auth H] HD [auth H] | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate C42 H81 O10 P ZEFGRNLJASLRBZ-QIJYXWHJSA-N |  | ||
| HEM (Subject of Investigation/LOI) Query on HEM | CD [auth C], DD [auth C], ED [auth C] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| SPN (Subject of Investigation/LOI) Query on SPN | AD [auth 7] BC [auth l] BE [auth 0] CB [auth w] FB [auth u] | SPEROIDENONE C41 H70 O2 GWQAMGYOEYXWJF-YCDPMLDASA-N |  | ||
| FE (Subject of Investigation/LOI) Query on FE | OD [auth M] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32370136 |