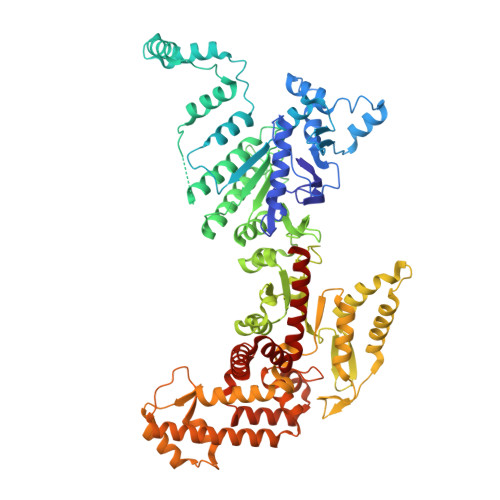
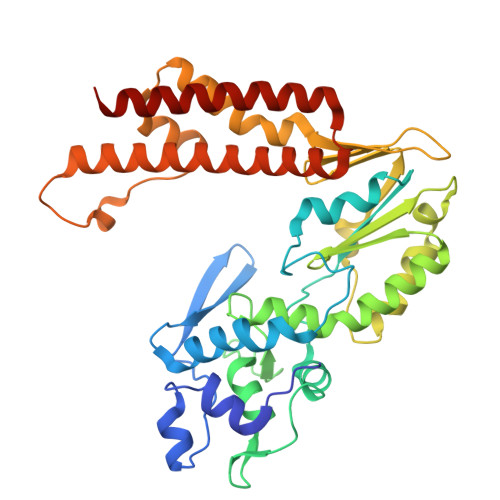
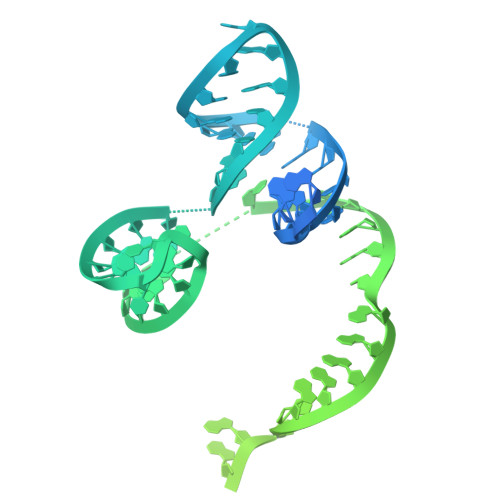
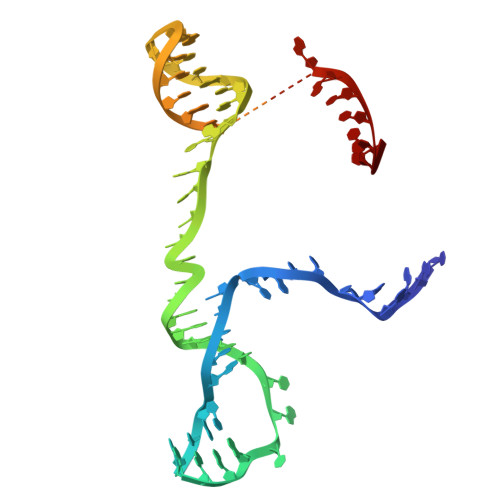
Molecular mechanism of Eco8-mediated anti-phage defense.
Yuan, L., Xu, L., Wu, B., Liu, Q., Yao, Y., Hua, X., Feng, Y.(2025) Mol Cell 85: 4229-4242.e4
- PubMed: 41172990
- DOI: https://doi.org/10.1016/j.molcel.2025.09.029
- Primary Citation of Related Structures:
9LP9, 9LPA - PubMed Abstract:
Escherichia coli Eco8 is an anti-phage defense system consisting of a reverse transcriptase, a class 3 overcoming lysogenization defect (OLD) nuclease, and a DNA-RNA chimera called multi-copy single-stranded DNA (msDNA). Genetic and biochemical data suggest that Eco8-mediated anti-phage defense is triggered by the phage single-stranded DNA (ssDNA)-binding proteins, but the underlying structural basis remains unknown. Here, we demonstrate that the DNA cleavage and ATP hydrolysis activities of the OLD nuclease are critical for Eco8-mediated anti-phage defense. We also determine the cryoelectron microscopy (cryo-EM) structures of Eco8 alone and in complex with the T7 phage ssDNA-binding protein. Structural analysis reveals that the reverse transcriptase, msDNA, and OLD nuclease form a megacomplex with a 4:4:4 stoichiometry. The T7 phage ssDNA-binding protein unwinds the msDNA and transforms Eco8 into an ATP-dependent DNA-degrading machinery. This study not only elucidates the molecular mechanism of Eco8-mediated anti-phage defense but also validates that msDNA serves as a sensor of phage DNA-modifying/binding proteins.
- Department of Infectious Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China; Department of Biophysics, Zhejiang University School of Medicine, Hangzhou, China.
Organizational Affiliation: